Review Article - (2023)Volume 9, Issue 2
New Insights into Porcine Endogenous Retrovirus (PERV)
Dong Niu1*, Tao Wang2, Yu Liu1 and Yifan Niu1Abstract
Xenotransplantation with porcine organs has been recognized as a promising solution to alleviate the shortage of organs for human transplantation. Porcine Endogenous Retrovirus (PERV), whose proviral DNAs are buried in the genome of all pig breeds, is a main microbiological risk for xenotransplantation. Over the last decades, some advances on PERVs’ study have been achieved. Here we reviewed the current progress of PERVs including the classification, molecular structure, regulation, function in immune system and potential risk in xenotransplantation. We also discussed the problem of insufficient research on PERVs as well as the questions need to be answered in the future work.
Keywords
Pig; Porcine Endogenous Retrovirus (PERV); Inflammation; Virus; Xenotransplantation; Epigenetic regulation
Introduction
With the development of regenerative medicine, transplantation has become the most effective solution for end stage organ failure. While the main difficulty in transplantation is the critical shortage of available donor organs. Xenotransplantation, which uses animals as the organ source for human transplantation, could potentially overcome the shortage of human organs. Pigs, which are anatomically and physiologically comparable to humans, are the most suitable donor animal for xenotransplantation [1-3].
However, three significant barriers need to be overcome before xenotransplantation becomes a clinical therapy. They are immune rejection, physiological incompatibility, and crossspecies transmission of Porcine Endogenous Retroviruses (PERVs) between the human recipient and the porcine xenograft.
To date, with the development of genome engineering and animal cloning technologies, it becomes possible to obtain multigene genetically modified donor pigs to overcome the immunological and physiological obstacles. And a recent study showed a baboon lived with a cardiac xenograft from a threegene (GTKO/hCD46/hTBM) modified pig survived beyond 900 days.
When analyzing the microbiological safety in xenotransplantation, PERVs pose the greatest threat. As an endogenous retrovirus, PERVs are integrated into the porcine genome and vertically transferred through inheritance, so PERVs cannot be eliminated by traditional ways such as Designated or Specified Pathogen-Free (DPF or SPF) breeding. In vitro experiments showed that PERVs could infect different human cells, and recombinant PERVs released by infected human cells acquired resistance to complement mediated killing. Like other retroviruses, PERVs can insert their cDNA into the host cell genome, potentially leading to immunodeficiency or tumorigenesis. Meanwhile, like most retrovirus, the transmembrane envelope protein of PERVs has a highly conserved immunosuppressive domain which can inhibit human immune cell function. This indicates that a high titer of PERV particles in the recipient who receives a solid vascularized porcine organ transplant might lead to an immunodeficiency disease. Meanwhile, there is no evidence from previous studies that PERVs can infect human recipients in vivo. However, most of these patients were not exposed to porcine grafts for a long time, and no immune suppressants were used as well [4-7].
Literature Review
Over the last decades, much progress has been made. However, a series of questions, such as what are the physiological functions of PERVs, and whether PERVs infect humans in vivo, still need to be answered in order to implement pig-to-human xenotransplantation.
This review presented the current knowledges on PERVs as well as the potential impact of PERVs on immune system and xenotransplantation.
Classification of PERVs
Endogenous Retrovirus (ERVs) discovery occurred in the late 1960’s and early 1970’s. The first reports on Avian Leucosis Virus (ALV), Murine Leukemia Virus (MLV), Murine Mammary Tumor Virus (MMTV) and PERVs were almost at the same time. Breese, et al. first described PERVs in 1970 as virus-like particles, same as those seen in the Baby Hamster Kidney (BHK-21) cell line infected with MLV. And according to the morphological properties, some investigators speculated that these viral particles belonged to the Type C Virus. In 1974, George, et al., further characterized the biochemical and immunological properties of the viral particles and demonstrated that it was a typical type C virus.
Two sets of human-tropic PERVs were first reported in 1997. By using the 3' Rapid Amplification of Cloned Ends (3'RACE), the Weiss team obtained two classes of sequences (PERV-A and PERV-B) from the PK15 and Human Embryonic Kidney 293 cells (HEK-293) infected with PERVs. The distinct third class of PERV is PERV-C, isolated from a swine lymphoma infected only with pig cells. The PERV A and B are found to be polytrophic, and the PERV C is ecotropic only infecting pig cells. In 2001, in addition to the PERV-A, B, and C, classified together as gamma genera of retroviruses (gamma-1), Patience, et al., detected the DNAs of four novel groups of gamma-retrovirus (gamma-2 to gamma-5) and four novel groups of beta-retrovirus (beta-1 to beta-4) in the genomes of domestic pigs. And so far, only the gamma-1 PERVs remain replication-competent as an active provirus [8-10].
PERV-A, B and C subfamilies are mainly different in Env gene sequence which encodes envelop protein, while the other genes encoding internal proteins, including gag and pol, are highly conserved among the three subfamilies. The PERV-A and PERVC could form a hybrid virus named as PERV-A/C through a recombination of the Env gene. As one part of the Env gene is originated from PERV-A, the recombinant PERV-A/C virus can infect human cells. And associated with alterations in the LTR sequences, the recombinant virus exhibits an increased replication capacity. Fortunately, by using PERV-C free pigs, the risk of PERV-A and PERV-C recombination in xenotransplantation can be eliminated.
Molecular structure of PERVs
PERVs are enveloped-linear ssRNA retroviruses containing two copies of the genomic RNA. Infectious PERVs (PERV-A, B, and C) belong to the Retroviridae family, Orthoretrovirinae subfamily, Gammaretrovirus genus. The transcript of PERVs has three Open Reading Frames (ORFs) which respectively encode structural proteins (Gag), Polymerases (Pol) and Glycoproteins (Env).
The Gag proteins consist of three major domains: The Matrix (MA), the Capsid (CA), and the Nucleocapsid (NC). The MA domain initially binds to the membranes and detaches from the host cell during budding.
The CA domain mediates the crystal lattice forming protein interactions in capsids. And the NC domain is responsible for the packaging of the viral RNA genome [11].
Discussion
In PERVs, the pol gene encodes the Protease (PR), the Integrase (IN), and the Reverse Transcriptase (RT). The PR can be autoactivated to cleave gag and pol at specific sites to initiate PERV maturation. The RT is a key enzyme in viral propagation, generating complementary DNAs (cDNAs) from the PERV ssRNA templates. With the help of the IN, the PERV cDNAs can be integrated into the host genome, from which the new viral copies can be generated via host transcription. The Env protein plays a critical role in the entry of PERV particles into the host cells during viral replication cycle. Env protein is synthesized as a trimeric precursor in the endoplasmic reticulum, which is subsequently cleaved into a surface Sub Unit (SU) and a Transmembrane subunit (TM) by a cellular furin-like PR. The SU subunit contains a Receptor-Binding Domain (RBD) which binds to a specific receptor on the target cell. Through the hydrophobic domain, the TM subunit is buried in the lipid bilayer of the viral particle [12]. And the cytoplasmic tail of the TM subunit contains an R peptide, which will be cleaved during virion maturation to activate the fusion capacity. The CxxC motif is localized at the C-terminus of the SU and covalently links the CXnCC motif on the TM to form an intersubunit disulfide bond. The disulfide bond anchors the SU subunit to the surface of PERV particles (Figure 1).
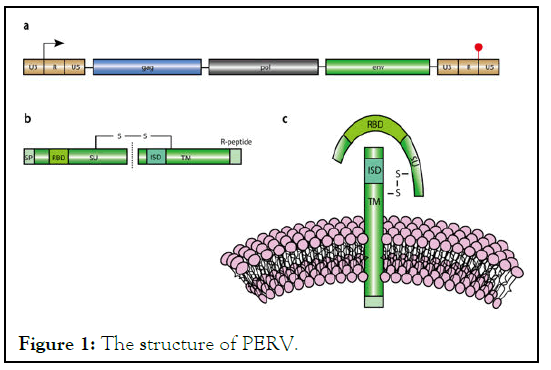
Figure 1: The structure of PERV.
In the proviral form of PERVs, the coding sequences of Gag, Pol, and Env are flanked by a 5' and a 3' Long Terminal Repeats (LTRs). And each LTR contains a Unique 3 (U3), Repeat region (R), and Unique 5 (U5) Regions in 5' to 3' direction: U3-R-U5. The transcription of PERVs starts at the U3-R junction in the 5ʹ LTR, and ends at the R-U5 junction in the 3ʹ LTR. LTRs play an essential role in the replication cycle of PERVs. Moreover, LTRs possess promoters, enhancers and multiple transcription factor binding sites, which drives the transcription of PERVs within the host genome.
Regulation of PERVs
Although in most normal healthy cells of pigs, the release of PERV particles (PERV-A, B, C) was not observed, PERVs were expressed in all tissues, with the highest expression in the lung, followed by the ovary, blood, liver and placenta. And a higher expression of PERV mRNA was found in melanomas than that in normal skin. It is of great interest that the integrated PERV proviral copy number differs among several organs of the same pig. As PERVs are transmitted vertically, like normal cellular genes, the number should be exactly the same in all organs of the same animal. These indicate that the expression and replication of PERVs are highly regulated both temporally and spatially during pig development [13].
LTRs play a vital role in the regulation of the PERVs within the host genome. In the LTR U3 region, there are directly repeated nucleotide sequences. A majority of these repeat boxes exist as one of the two types, which are distinguished by a 39 bp (PERVA and B) or 37 bp (PERV-C) of the repeated nucleotides. And the multiplications of these repeat boxes also correlate with relatively late viral infection event, higher viral production, and higher infectious viral titer. After PERVs integrate into the host genome, these repeat boxes may increase the transcription of viral genes and flanked cellular genes. In response to the deleterious effects of the replicating PERVs, the number of these repeat boxes is restricted by natural instability and the constraints imposed by virion packaging limits [14].
Heritable changes in PERV expression without changing the DNA sequence can occur through altering DNA methylation or histone modification. In mice, the Endogenous Retroviruses (ERVs) cause an estimated 10% of germline mutations in current inbred laboratory strains. Transcription of ERVs is generally repressed by epigenetic marks such as DNA methylation and histone modification. Likewise, treatment of cells with 5‑azacytidine, which promotes the demethylation of genomic DNA, results in PERV induction. It indicates the transcription of PERVs is sensitive to DNA methylation, and most PERV proviruses in pig genomes are currently silenced by DNA methylation on PERV 5' LTRs CpG Island. And the PK-15 cell line, which has a very low DNA methylation level, keeps a high PERV transcriptional profile. And viral production also can exemplify the relation between the DNA methylation and the PERV mRNA transcription. Transcriptional silencing can also be achieved via establishment of a heterochromatic structure, which maintains an inactive form of chromatin [15].
The highly condensed conformation prevents the access of transcriptional machinery and eventually suppresses transcriptional expression. Heterochromatin formation is associated with several histone marks in the genome, such as trimethylation of H3K9 (H3K9me3) and H4K20 (H4K20me3). Primarily the H3K9me3 has been found to be involved in inactivating viral chromatin of ERVs. Meanwhile, knock-out experiments of methyltransferase, which generates the H3K9me3, have shed light on their involvement in ERVs silencing.
Likewise, PERVs are marked by the H3K9me3 and H4K20me3, and these marks were not restricted in porcine embryonic germ cells (pEGCs), but also apparent in PK15 cells. Interestingly, the younger and more active PERV subfamilies are generally marked by higher histone methylation levels than the ancient inactivated PERV subtypes [16].
The role of PERVs in immune system
Multiple immune stimuli have been shown to induce ERV expression. In mice, Lipo Poly Saccharides (LPS), Lipo Proteins (LP) and 5-Bromo-2’-Deoxyuridine (BrdU) were able to induce ERV expression. ERV assessment in gut tissues from different conditions revealed that expression of certain ERVs depended on the presence of the gut microbiota. Recently, it was shown that the host immune system could use the ERVs to communicate with the exogenous microbiota that modulated tissue repair or inflammation. At the same time, an increased expression of PERVs was found in mitogen-stimulated porcine lymphocytes, indicating that immune stimulation may also activate PERV expression in vivo [17].
Most exogenous retroviruses have been described to suppress the host's immune system. By analyzing the mechanism of immunosuppression by retroviruses, it was shown that the Env protein has a highly conserved so-called Immuno Suppressive Domain (ISD) in the transmembrane unit that is responsible for the immunosuppressive activity. Exposure of Peripheral Blood Mononuclear Cells (PBMCs) to ISD derived peptides increased the expression of different cytokines meanwhile inhibited mitogen-driven cell proliferation. PERVs remain the intact Env proteins and the ISD related domain as a molecular relic of the ancient retroviruses. It was shown that purified inactivated- PERV virus and the ISD related peptide inhibited proliferation of human PBMCs and induced IL-10 production in PBMCs.
The impact of ERVs on the immune response is very complicated. On the one hand, the conserved ISD domain in the Env protein inhibits the host innate immune system. On the other hand, ERVs' replication intermediates can stimulate the innate sensors and trigger the Interferons (IFNs) response.
In the innate immune system, the Pattern Recognition Receptors (PRRs) are key components that can directly recognize Pathogen Associated Molecular Patterns (PAMPs) and trigger the innate immune defence line. There are two main families responsible for recognizing the nucleic acids of ERVs. One is Toll Like Receptor (TLR) family which monitors the contents of endolysosomal compartments. The other is cytosolic PRRs recognizing nucleic acids in the cytoplasm. The PRRs include the RIG-I-Like Receptors (RLRs) and analogous DNA sensing receptors such as cGAMP Synthase (cGAS), etc. Once ERVs are activated by stimuli like 5-Azacytidine (AZA), which promotes ERVs expression, PRRs are triggered following signal cascades and lead to type I IFN production. In the same way, the activation of ERVs could stimulate anti-tumor immunity by promoting the viral recognition and interferon response pathway. However, the association between PERVs and the porcine immune system remains poorly understood. Interestingly, there was an increased expression of PERVs in the tumors compared with that in the normal pig cells. The PERV viral particles had been isolated from lymphoma cells, radiation-induced leukemia cells and PK-15 cell lines. It indicated PERVs might play a beneficial role in the porcine innate immune system [18].
Potential risk of PERVs in xenotransplantation
PERV-A and PERV-B are found to be able to infect various kinds of human cells in vitro, therefore PERVs represent a potential risk for xenotransplantation. Although there has been no report of PERV transmission in clinical trials, some rare cases like antibodies against PERV structural proteins have been found, indicating the impact of PERVs in xenotransplantation need to be further explored.
Like other retroviruses, PERVs are able to integrate their viral cDNA into the genome of the host cell. As the result of random insertion, PERVs may destroy the normal genes and irreversibly break the balance between the expression of oncogenes and tumor suppressor genes. Some in vitro studies had shown that PERVs-infected human cells were able to produce LTR sequence-altered PERVs, and these mutated viruses not only increased reverse transcriptional activity but also enhanced the infectivity. Moreover, Patience, et al., found that PERVs released by the infected human cells were resistant to complementmediated lysis, further demonstrating the potential risk of PERVs in xenotransplantation [19].
Human Endogenous Retroviruses (HERVs) account for 5%-8% of the human genome. Same as PERVs, HERVs are remnant relics of the exogenous retroviruses in ancient germline infections. Although most HERVs are inactivated after million years of evolution, several of these proviruses have retained partial coding capacity. And many recent studies have shown that HERVs are not only related to cancer and neurodegenerative diseases, but also involved in shaping cell types and chromosome structures. In addition, HERV-K related proteins participate in the immune pathway in the early stage of human embryonic development. In the human genome, there are a large number of gamma-retroviruses, such as HERV-T, HERV-E and HERV-I, that are closely related to PERVs. Once the recombination's between PERVs and HERVs occur, it might possibly result in a recombinant pathogenic strain, posing a new threat to human society.
Several studies have demonstrated that transcripts and proteins of HERVs are up-regulated in patients with retrovirus HIV-1 infections. It has been reported that the HIV-1 Tat protein is able to activate the HERV-K LTRs directly. Whether PERVs would influence HERVs’ expression like HIV still remains unknown. Using the CRISPR system targeting the catalytic core of RT enzyme of PERVs, we successfully produced PERVs inactivated PK-15 cells and live pigs. Caused by the RT knockout, these virions were no longer infectious, and the usage of the PERVs inactivated donor pigs in xenotransplantation would eliminate most threats caused by the random integration of PERV proviral cDNAs into host genome. Recent studies have further checked our RT-inactivated PK-15 cells, and confirmed the PERVs released by the cells were impaired non-infectious viral particles. However, due to the intact sequence of PERV Env and Gag genes, the RT-inactivated PERVs still carry normal viral proteins such as Env protein which is required for viral adherence and uptake. It raises new questions that whether the intact Env or Gag protein would upregulate the expression of HERVs and trigger a negative influence on the recipients [20].
Conclusion
Since the risk of PERV infection is only amplified in xenotransplantation, the studies of PERVs have been proceeding slowly in the past decades. With the advent of the CRISPR technology, genetic modification can overcome immune rejection, physiological disorders and other obstacles in xenotransplantation. Meanwhile, understanding the role of PERVs in the porcine immune system will be helpful in revealing the pathogenesis of porcine diseases and alerting the risk in xenotransplantation. The mechanisms of how PERVs are regulated by the host and how they interact with HERVs in xenotransplantation require more attention and further exploration. Moreover, the deficiency in PERV studies will become a barrier to hinder the progress of xenotransplantation, and thus more efforts need to be taken on this pivotal subject.
Ethics Declaration
No ethical approval was required as this is a review article with no original research data.
Conflict Of Interest Statement
All authors have no competing interests.
Funding
This work was supported by the key project of natural science foundation of zhejiang province (LZ22C010003), the fundamental research funds for the provincial universities of zhejiang (2020YQ007) and Zhejiang A and F university talent initiative project (Nos: 2019FR022, 2019FR052 and 2020FR033).
Authors' Contribution
Dong Niu conceived and supervised the work. Yu Liu and Yifan Niu drafted the manuscript. Tao Wang drew the figures.
References
- de Akiyoshi, Denaro M, Zhu H, Greenstein JL, Banerjee P, Fishman JA. Identification of a full length cDNA for an endogenous retrovirus of miniature swine. J Virol. 1998;72(5):4503-4507.
[Crossref] [Google Scholar] [PubMed]
- Blusch JH, Seelmeir S, von der Helm K. Molecular and enzymatic characterization of the porcine endogenous retrovirus protease. J Virol. 2002;76(15):7913-7917.
[Crossref] [Google Scholar] [PubMed]
- Bobkova M, Stitz J, Engelstadter M, Cichutek K, Buchholz CJ. Identification of R-peptides in envelope proteins of C-type retroviruses. J Gen. Virol. 2002;83(9):2241-2246.
[Crossref] [Google Scholar] [PubMed]
- Breese SS. Virus like particles occurring in cultures of stable pig kidney cell lines. Arch Gesamte Virusforsch. 1970;30:401-404.
[Crossref] [Google Scholar] [PubMed]
- Chiappinelli KB, Zahnow CA, Ahuja N, Baylin SB. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 2016;76(7):1683-1689.
[Crossref] [Google Scholar] [PubMed]
- Cianciolo GJ, Copeland TD, Oroszlan S, Snyderman R. Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Sci. 1985;25;230(4724):453-455.
[Crossref] [Google Scholar] [PubMed]
- Denner J. Recombinant porcine endogenous retroviruses (PERV-A/C): A new risk for xenotransplantation?. Arch Virol. 2008;153:1421-1426.
[Crossref] [Google Scholar] [PubMed]
- Denner J. The transmembrane proteins contribute to immunodeficiencies induced by HIV-1 and other retroviruses. Aids. 2014;28(8),1081-1090.
[Crossref] [Google Scholar] [PubMed]
- Denner J. How active are Porcine Endogenous Retroviruses (PERVs)?. Viruses. 2016;8(8):215.
[Crossref] [Google Scholar] [PubMed]
- Dieckhoff B, Puhlmann J, Buscher K, Hafner-Marx A, Herbach N, Bannert N, et al. Expression of Porcine Endogenous Retroviruses (PERVs) in melanomas of Munich miniature swine (MMS) Troll. Vet. Microbiol. 2007;123(1-3):53-68.
[Crossref] [Google Scholar] [PubMed]
- Dinsmore JH, Manhart C, Raineri R, Jacoby DB, Moore A. No evidence for infection of human cells with Porcine Endogenous Retrovirus (PERV) after exposure to porcine fetal neuronal cells. Transplant. 2000;70(9):1382-1389.
[Crossref] [Google Scholar] [PubMed]
- Fiebig U, Fischer K, Bahr A, Runge C, Schnieke A, Wolf E, et al. Porcine endogenous retroviruses: Quantification of the copy number in cell lines, pig breeds, and organs. XenoTransplant. 2018;25(4):e12445.
[Crossref] [Google Scholar] [PubMed]
- Godehardt AW, Fischer N, Rauch P, Gulich B, Boller K, Church GM, et al. Characterization of porcine endogenous retrovirus particles released by the CRISPR/Cas9 inactivated cell line PK15 clone 15. XenoTransplant. 2020;27(2):e12563
[Crossref] [Google Scholar] [PubMed]
- Groh S, Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci. 2017;74(11):2055-2065.
[Crossref] [Google Scholar] [PubMed]
- Guell M, Niu D, Kan Y, George H, Wang T, Lee IH, et al. PERV inactivation is necessary to guarantee absence of pig to patient PERVs transmission in xenotransplantation. XenoTransplant. 2017;24(6):e12366.
[Crossref] [Google Scholar] [PubMed]
- Armstrong JA, Porterfield JS, de Madrid AT. C-type virus particles in pig kidney cell lines. J Gen Virol. 1971;10(2):195-198.
- Barbalat R, Ewald SE, Mouchess, ML, Barton, GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011.
- Pornillos O, Ganser-Pornillos BK. Maturation of retroviruses. Curr Opin Virol. 2019;36:47-55.
[Crossref] [Google Scholar] [PubMed]
- Roulois D, Yau HL, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961-973.
[Crossref] [Google Scholar] [PubMed]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. LTR-retrotransposon control by tRNA derived small RNAs. Cell. 2017;170(1):61-71.
[Crossref] [Google Scholar] [PubMed]
Author Info
Dong Niu1*, Tao Wang2, Yu Liu1 and Yifan Niu12Department of Genetics, University of Genetic Engineering, Nanjing, China
Citation: Niu D, Wang T, Niu Y, Liu Y (2023) New Insights into Porcine Endogenous Retrovirus (PERV). Appl Microbiol. 9:247.
Received: 28-Nov-2022, Manuscript No. AMOA-22-20487; Editor assigned: 30-Nov-2022, Pre QC No. AMOA-22-20487 (PQ); Reviewed: 07-Dec-2022, QC No. AMOA-22-20487; Revised: 17-Feb-2023, Manuscript No. AMOA-22-20487 (R); Published: 24-Feb-2023 , DOI: 10.35284/2471-9315.23.9.247
Copyright: © 2023 Niu D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.