Journal of Medical Diagnostic Methods
Open Access
ISSN: 2168-9784
ISSN: 2168-9784
Research Article - (2022)Volume 11, Issue 4
3M syndrome is a disorder that origins the skeletal abnormalities including short stature (dwarfism) and unusual facial features. The name of this condition comes from the initials of three researchers who first identified it: Miller, McKusick, and Malvaux. Individuals with 3M syndrome grow extremely slowly before birth, and this slow growth continues throughout childhood and adolescence. They have low birth weight and length and remain much smaller than others in their family, growing to an adult height of approximately 4 feet to 4 feet 6 inches (120 centimeters to 130 centimeters). In this study we are reporting a novel mutation of 3M Syndrome in an Indian subject. We have performed exome sequencing and after identification of likely pathogenic variant we have validated this mutation with Sanger sequencing method. Further research is rational to regulate the classic and the variant presentations of this condition, with the follow-up of the patients that provides the valuable data insights into its natural history and the long-term prognosis. As per database these variants have been not reported till date, we found a novel variant in an Indian patient. This study highlights the clinical utility of the exome sequencing in the definite diagnosis of provisionally diagnosed genetic disorders.
Exome sequencing; 3M Syndrome; Skeletal abnormalities; Pre and post-natal growth
3M syndrome is a disorder that causes the skeletal aberrations with short height and the abnormal facial appearance. The name of this condition comes from the identifies of three researchers who first acknowledged the name 3M syndrome (Miller, McKusick, and Malvaux). The mutations in any 1 of the subsequent 3 genes CUL7, OBSL1, and CCDC8 are blamable for the manifestation of this disorder [1]. The inheritance pattern of this disorder is an autosomal recessive and is deliberated very rare, so far less than 100 cases worldwide have been identified till date. In year 2005, the autozygosity mapping discovered a 3M syndrome locus on the chromosome 6p21.1, and the pathogenic mutations in the CULLIN7 (CUL7 (MIM 609577)) were subsequently recognized as the primary cause of 3M syndrome. More newly, further the autozygosity mapping discovered a second 3M syndrome locus on the chromosome 2q35-q36.1 with the basic mutations identified in Obscurin-like 1.
The people with 3M syndrome agonize from the severe prenatal growth obstruction owing to growth adjournments during the fetal development, resultant in a low birth weight. They have the low birth weight and length and remain much smaller than others in their family, growing to an adult height of approximately 4 feet to 4 feet 6 inches (120 centimeters to 130 centimeters). In some affected individuals, the head is normalsized but looks disproportionately large in comparison with the body. In other people with this disorder, the head has an unusually long and narrow shape (dolichocephaly). Intelligence is unaffected by 3M syndrome, and life expectancy is generally normal. The growth adjournments continue after birth throughout childhood and adolescence, ultimately leading to a short stature. Many of the physical features associated with the disorder are congenital. The distinguishing craniofacial abnormalities typically comprise a long, narrow head that is inconsistent to the body size, a broad and the prominent forehead, and a triangular-shaped face with a hypoplastic midface, long philtrum, pointed chin, depressed nasal bridge, fleshy-tipped reversed nose, protruding mouth, large ears, and the full lips [2-4].
Other skeletal irregularities that repeatedly occur in this disorder comprise a short, broad neck and chest; prominent shoulder blades; and shoulders that slope less than usual (square shoulders). The affected individuals may have abnormal spinal curvature such as a rounded upper back that also curves to the side (kyphoscoliosis) or inflated curvature of the lower back (hyperlordosis). People with 3M syndrome can also have the unusual curving of the fingers also called clinodactyly, short pinky (small) fingers, prominent heels, and the loose joints. The additional skeletal irregularities, such as remarkably slender long bones in the arms and the legs; tall, narrow spinal bones (vertebrae); or the slightly delayed bone age may be apparent in the x-ray pictures.
A variant of 3M syndrome called the Yakut short stature syndrome has been acknowledged in the isolated Yakut population in the Russian province of Siberia. In adding to having most of the physical features characteristic of 3M syndrome, people with this form of the disorder are often born with breathing problems that can be life-threatening in infancy. The prevalence of 3M syndrome is unknown. About 100 individuals worldwide with this disorder have been described in the medical literature.
The genetic background of 3M syndrome
Mutations in the CUL7 gene causes 3M syndrome in more than three-quarters of affected individuals, including those in the yakut population. The mutations in the OBSL1 gene cause around 16 percent of cases of this disorder. Mutations in other genes, some of which have not been identified, account for the remaining cases.
The CUL7 gene provides the instructions for making a protein called cullin-7. This cullin-7 protein shows a role in the cell machinery that breakdown or degrades the unwanted proteins, called a system named ubiquitin-proteasome system. Specifically, the cullin-7 helps in the assembling a complex called an E3 ubiquitin ligase, which tags the unneeded proteins for the degradation. The protein produced from the OBSL1 gene is thought to help maintain normal levels of the cullin-7.
The ubiquitin-proteasome system helps in the regulation of the level of proteins tangled in several critical cell activities such as the timing of cell division and the growth. In particular, the proteins formed from the genes related with 3M syndrome are thought to help into regulate the proteins involved in the body's response to the growth hormones, although their specific role in this process is unknown [5].
Mutations in the CUL7 or OBSL1 gene prevent the cullin-7 protein from bringing together the components of the E3 ubiquitin ligase complex, interfering with the process of tagging unneeded proteins for degradation. The body's response to growth hormones may be impaired as a result. However, the specific relationship between CUL7 and OBSL1 gene mutations and the signs and symptoms of 3M syndrome are unknown [6].
Patient information: A 7 months old female patient was presented to the clinician with the family history of the shortening of the limbs, the abnormal ribs and the cleft palate. Sibling has similar condition in the family history (Table 1).
| Gene | Chromosomal Coordinates | Exon | Variant | Zygosity | Condition group | Significance | Inheritance | Coverage |
|---|---|---|---|---|---|---|---|---|
| CUL7 | chr6:43019009:GG CTTGCACCA:G NM_001168370.1 | 4 | c.1172_1181 delTGGTGCA AGC p.Leu391fs | Homozygous | 3M syndrome 1 | Likely pathogenic | Autosomal recessive | 88 |
Table 1: Findings of after the accomplishment of exome sequencing.
DNA extraction and quality analysis: The genomic DNA was extracted from the 2 mL of peripheral venous blood samples of the patient, with DNA extracted by using the standard phenol– chloroform method [7]. The quality of DNA was measured on the 2% agarose gel electrophoresis, and the quantity of the DNA was measured by the nano drop TM.
The exome sequencing: The DNA extracted from blood was used to perform the targeted gene capture using an agilent sure select exome V6 kit. The libraries were sequenced to mean and gt; 100 X coverage on illumine sequencing platform and 6 to 8 GB data will be generated. The sequences found are aligned to human reference genome (GRCh37/hg19) and variant analysis was performed using set of bioinformatics pipeline. Clinically relevant mutations were annotated using published variants in literature and a set of diseases databases–ClinVar, OMIM, GWAS, HGMD and SwissVar. Common variants are filtered based on allele frequency in 1000 Genome Phase 3, ExAC, EVS, dbSNP147, 1000 Japanese Genome etc. Non-synonymous variants effect is calculated by using the multiple algorithms such as polyphen-2, SIFT, mutationtaster 2, mutation assessor and LRT. Only non-synonymous and the splice site variants found in the clinical exome panel comprising of the specific set of the genes were used for clinical clarification. Silent variations that do not result in any change in amino acid in the coding region are not reported.
Sanger validation: For the confirmation of novel likely pathogenic mutation identified in Exome sequencing, we have validated it by gold standard technique sanger sequencing.
After performing the exome sequencing in the patient. The individual carries two copies (homozygous) of a frame shift variant in CUL7 gene which is predicted to cause a frame shift and consequent premature truncation of the protein. The variant has been reported in dbSNP database with an identification number rs1160826185 and in the genome aggregation database (gnomAD) with a rare allele frequency of 0.0003990%. Since, this variant is predicted to produce a truncated protein which might result in loss of function and other frame shift truncating variants in this gene are also known to cause similar phenotype, therefore, this variant has been labeled as likely pathogenic.
This variant is responsible for 3M syndrome is an autosomal recessive disorder characterized by distinctive facial features, severe prenatal and postnatal growth retardation, and normal mental development. The main skeletal anomalies are long, slender tubular bones, reduced anteroposterior diameter of the vertebral bodies, and delayed bone age. Other skeletal manifestations include joint hypermobility, joint dislocation, winged scapulae and pes planus.
The protein encoded by this gene is a component of an E3 ubiquitin-protein ligase complex. The encoded protein interacts with TP53, CUL9, and FBXW8 proteins. Defects in this gene are a cause of 3M syndrome type 1 (3 M 1). Two transcript variants encoding different isoforms have been found for this gene. The Sanger sequencing confirmation of the variant in the CUL7 gene was accomplished (Figure 1).
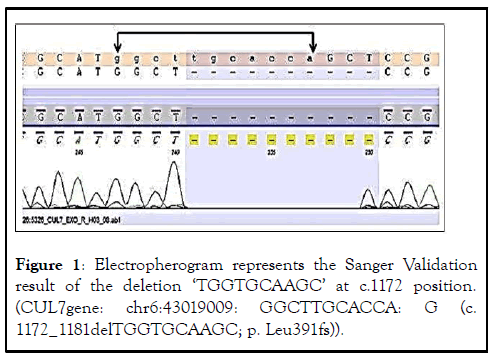
Figure 1: Electropherogram represents the Sanger Validation result of the deletion ‘TGGTGCAAGC’ at c.1172 position. (CUL7gene: chr6:43019009: GGCTTGCACCA: G (c. 1172_1181delTGGTGCAAGC; p. Leu391fs)).
3M syndrome is categorized by a triangular-shaped face with the frontal bossing, sunken nasal bridge, and mild malar hypoplasia with a plump tip of nose, full lips and upturned nares. The patients classically have larger heads for their height, the dolichocephaly, and the normal intelligence. The other clinical findings are a short wide thorax, brachydactyly, clinodactyly, micromelia, and the prominent heels. Both the sexes are affected likewise. There no hormonal deficiencies are detected. Our patient had nearly all of the typical clinical features, along with the above-mentioned bony abnormalities, but did not show radiological findings of slender long bones with thin diaphysis and the tall vertebral bodies. These findings may appear future in some patients. The radiographic examination, although abnormal, is not diagnostic, as similar X-ray changes have been documented in other disorders.
In the study of stated on 43 patients with the short stature, hydrocephaloid skull, and the typical face in an isolated Yakut population. Although the clinical outcomes were similar to the 3M syndrome but the slender long bones and the tall vertebral bodies have not been usually observed in this short-stature syndrome in [8], as in our patient [9].
Another study done by described the 9 cases of children with the primordial dwarfism and the facial dysmorphism regarded as the ‘gloomy face.’ Despite very short stature, there were no any radiologic irregularities of the bones and there was no hormone deficiency was present. Recommended that this was a distinct complaint with the features of 3M syndrome [10].
The Physical findings of the numerous individuals such as Silver- Russell Syndrome (SRS) are parallel to the 3M syndrome. Silver- Russell syndrome has many similarities with 3M syndrome: intrauterine growth retardation, short stature, triangular face, relatively large skull, asymmetry of body or limbs, and the clinodactyly. Mild mental retardation also can be found in patients with SRS, but abnormalities of the skeletal system have not been reported.
By using the direct sequencing method the reported 25 dissimilar mutations in the cullin-7 gene in 29 families with 3M syndrome [11-13].
In all 43 affected individuals with Yakut short-stature syndrome, identified homozygosity for a founder mutation in the CUL7 gene. Delayed bone age has been reported in 3M syndrome patients; our patient’s was 1 year behind his chronological age. An early diagnosis is very significant for the genetic counseling in the 3M syndrome, particularly in different Muslim countries, where the consanguineous marriages are very common, and the autosomal recessive genetic disorders leading to severe short stature should be kept in mind. 3M syndrome should always be considered in the differential diagnosis of patients with growth retardation of prenatal onset.
In conclusion this study showed the utility of exome sequencing in misdiagnosed pathogenic cases. By this study we reported a novel mutation in the Indian subcontinent. Exome approach provides a major advance in the likely diagnostic yield in suspected inherited novel mutation containing cases; this methodology may uncover unexpected findings such as mutations in genes known to cause familial.
PV drafted the manuscript. AD reviewed all the study. DK gave patients details and VKM provided the bioinformatics inputs in whole exome sequencing data. All authors contributed to the article and approved the submitted version.
The authors would like to thank all the study participants. We are grateful to the physicians who referred patients for the further investigation.
Informed consent was obtained from all subjects involved in the study.
Citation: Vishwakarma P, Dubey A, Kalo D, Mishra VK (2022) Novel Mutation Documentation of 3M syndrome by Exome Sequencing in an Indian Subject. J Med Diagn Meth. 11:380.
Received: 19-May-2022, Manuscript No. JMDM-22-17585; Editor assigned: 20-May-2022, Pre QC No. JMDM-22-17585(PQ); Reviewed: 03-Jun-2022, QC No. JMDM-22-17585; Revised: 16-Sep-2022, Manuscript No. JMDM-22-17585(R); Published: 26-Sep-2022 , DOI: 10.35248/2168-9784.22.11.380
Copyright: © 2022 Vishwakarma P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.