
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research Article - (2024)Volume 7, Issue 1
This study aimed to assess in vitro the impacts of Nuclear Factor E2-Related Factor 2 (NRF2) knockdown on the transformation of adipose phenotype and the possible mechanisms of resistance to aging in 3T3-L1 cells. In the current study, the Nrf2-Knockdown (NK) via siRNA transfection increased the expression of Brown Adipose Tissue (BAT) marker genes including Peroxisome Proliferator-Activated Receptor-Gamma Coactivator (PGC)- 1α, Type 2 Iodothyronine Deiodinase (DIO2) and Positive Regulatory Domain Zinc Finger Region Protein 16 (PRDM16) and lowered the gene and protein expression of White Adipose Tissue (WAT) marker genes for instance Bone Morphogenetic Protein 4 (BMP4), Resistin and Retinoblastoma 1 (Rb1) in adipocytes; NK also altered the protein expression of longevity-related genes, such as Sirtuin 1 (Sirt1) and AMP-activated protein kinase (AMPK ) and Increased Mitochondrial Uncoupling Protein (UCP1) and Cytochrome C (CYCS), which are involved in mitochondrial generation. These results support the potential of Nrf2 as a possible therapeutic target for delaying aging through the transformation of adipose-phenotype and the effect of longevity factors.
Nuclear factor E2-related factor 2; 3T3-L1 cells; Adipose; Adipose-phenotype; Longevity
With the development of society, aging and obesity have become the major social problems in the world, especially in China [1]. One of the basic processes related to aging is the imbalance in the regulation of energy homeostasis [2]. Many age-related genes belong to evolutionarily conserved pathways that also control energy metabolism and Adipose Tissue (AT) plays an important metabolic role in regulating energy homeostasis throughout the body [3]. AT has long been thought of as an organ that stores and releases energy [4]. However, studies in recent years have shown that AT is also the largest endocrine tissue in human body. Through adipocytes and macrophages, a series of hormones, adipokines, cytokines and inflammatory factors are secreted to regulate physiological activities in human body, including diet, glucose, lipid and energy metabolism [5,6]. AT is now known to participate in a broad spectrum of inter-organ communication [7]. However, the specific function of AT and its effect on aging remain largely unknown. The role of AT in aging and longevity has become the focus of medical research.
AT in the body is mainly divided into Brown Adipose Tissue (BAT) and White Adipose Tissue (WAT). With individual growth and development, BAT content gradually decreases [8]. Recent studies have found that BAT is not only essential for metabolism, but also plays an important role in delaying aging and other processes [9]. Some studies have shown an inverse correlation between age and the metabolic activity of BAT. Notably, many of the beneficial consequences of BAT activity overlap with metabolic biomarkers of extended lifespan and health span. The most readily observed changes in BAT during aging in humans and mice are the loss of tissue mass and activity [10]. Therefore, there is growing interest in the potential role of BAT in delaying the aging process. WAT is a major fat component and a major storage site for body fat. WAT is also an activated endocrine organ that affects homeostasis throughout the body. It increases pro- inflammatory factors, leading to fat deposition in surrounding organs and insulin resistance, which is a key pathological process of obesity-related diseases [11,12]. These may be important reasons for the increased harmfulness of WAT during aging.
Nuclear factor erythroid-2-related factor is an important anti- oxidative stress transcription factor in the body, it regulates the expression of various detoxifying enzymes [13]. According to the theory that oxidative free radicals cause aging, Nrf2 is not only an antioxidant and detoxification reaction. Recent studies have shown that Nrf2 also plays an important role in fat metabolism and glucose metabolism [14], but whether Nrf2 has a sub induced effect on the transformation of adipose phenotype and its effect on aging is still less clear. Therefore, in this study, we explored the effect of Nrf2-knockdown on adipose-phenotype and longevity-related factors expression in adipocytes in vitro.
Cell culture and adipocyte differentiation
Murine 3T3-L1 preadipocytes purchased from the cell bank, Chinese Academy of Sciences were maintained in DMEM supplemented with 10% nerborn calf serum. Briefly, two-day post confluent cells were treated with 0.5 mM Methylisobutylxanthine, 1 μm Dexamethasone (D), 1.7 μM Insulin (I). Mix well and feed cells to initiate differentiation. The cells were left in the Methylene Diphenyl Diisocyanate (MDI) mix for 48 h. At 48 hours, exchange media for Dulbecco′s Modified Eagle′s Medium (DMEM) high glucose, 10% Fasting Blood Sugar (FBS) and 1/4 original concentration of insulin. Add the stock solution of insulin (1 mg/ml, filter sterilized at induction) at 0.25 ml per 100 ml media. Exchange media every 2 days thereafter using DMEM high glucose, 10% FBS and pen/strep [4].
Knockdown of Nrf2 expression by siRNA
The small-interfering Ribonucleic Acid (siRNA) utilized in this study targeted the following sequences of the mouse Nrf2gene: sense, 5’-GAAUUACAGUGUCUUAAUATT-3’ and antisense, 5’-UAUUAAGACACUGUAAUUCTT-3’. The Nrf2 siRNA were purchased from sigma. The DharmaFECT Duo Transfection Reagent was purchased from the Thermo Fisher Scientific (Waltham, MA), according to the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction analysis
After siRNA transfection, 3T3-L1 adipose cells were collected for total RNA extraction. Total RNA from each group was isolated using a Hipure Total RNA Mini Kit (Cat.#: R4111-03, Magen, Guangzhou, China). cDNA was synthesized from 2 μg of extracted total RNA using a ReverTra Ace qPCR RT kit (Cat.#: FSQ-101, TOYOBO, Kita-ku, Osaka, JAPAN). The primer sequences used in this study are shown in Table 1.
| Gene | Forward primer (5'-3') | Reverse primer (3'-5') |
|---|---|---|
| Nrf2 | CTACAGTCCCAGCAGGACATGGATTTG | GTTTTCGGTATTAAGACACTGTAATTCGGGAATGG |
| HO-1 | CCCCACCAAGTTCAAACAGC | AGCTCCTCAAACAGCTCAATGT |
| NQO1 | AGGATGGGAGGTACTCGAATC | TGCTAGAGATGACTCGGAAGG |
| Sirt1 | CATTTATCAGAGTTGCCACCAA | ACCAACAGCCTTAAAATCTGGA |
| PGC-1α | CCGAGAATTCATGGAGCAAT | GTGTGAGGAGGGTCATCGTT |
| UCP1 | AATGGTGAAGGTCGGTGT | TGGAGTCATACTGGAACATGTAG |
| Resistin | CAGGACCTGTATGCTTTAGGATG | TGTCCAGTCTATCCTTGCACAC |
| Rb1 | CAAATTAGAACGGACGTGTGAAC | ACCAGGTCATCTTCCATCTGT |
| Cycs | AATCTCCACGGTCTGTTCGG | TCTCCCCAGGTGATGCCTTT |
| Dio2 | CAAACAGGTTAAACTGGGTGAAGA | GTCAAGAAGGTGGCATTCGG |
| PRDM16 | AGGAGGAAGATCCAGGGGAC | GCCCATCCAAGCAGAGAAGT |
| BMP4 | GCCATTCACTATACGTGGACTTC | CAGAGTTGACTAGGGTCTGCAC |
| AMPKα | GGGTGAAGATCGGCCACTAC | TGCTTGCCCACCTTCACTTT |
| Gapdh | AATGGTGAAGGTCGGTGT | TGGAGTCATACTGGAACATGTAG |
Table 1: A list of PCR primers used for this study.
Western blot analysis
Protein lysates were resolved by gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membrane was blocked for 1 h in 5% nonfat milk, subsequently probed with specific antibodies and visualized using a ChemiDocTM MP system (Bio-Rad, Hercules, CA). GAPDH was used as the loading control. Protein band density was quantified using ImageJ software and normalized to that of Gapdh and β-tubulin (antibodies listed in Table 2).
| Antibody | Company | Reference | Dilution |
|---|---|---|---|
| Nrf2 | Santa cruz | Sc-722 | 0.736111111 |
| HO-1 | Abcam | ab52947 | 0.736111111 |
| NQO1 | Proteintech | 11451-1-AP | 0.736111111 |
| AMPKα | Ruiying biological | RLT0215 | 0.736111111 |
| Sirt1 | Cell signaling | Q96E86 | 0.736111111 |
| PGC-1α | Abcam | ab54481 | 0.736111111 |
| UCP1 | Abcam | ab23841 | 0.736111111 |
| Resistin | Abcam | ab216840 | 0.319444444 |
| Rb1 | Santa cruz | Sc-73598 | 0.736111111 |
| Cycs | Abcam | ab2757022 | 0.736111111 |
| Dio2 | Abcam | ab77779 | 1.430555556 |
| PRDM16 | Abcam | ab106410 | 0.736111111 |
| BMP4 | Abcam | ab39973 | 0.736111111 |
| Gapdh | Santa cruz | Sc420485 | 0.736111111 |
| β-tubulin | Santa cruz | Sc-5274 | 0.736111111 |
Table 2: Antibodies used for Western blot analysis for this study.
The morphology of 3T3-L1 cells induced by MDI and results of siRNA transfection
The 3T3-L1 cells cultured in 10% calf serum/DMEM grew to 100% confluence (Figure 1A). The full differentiation of 3T3- L1 adipocytes was usually achieved by day 8 (Figure 1B). Nrf2 knockdown after 24 h of siRNA transfection was confirmed by Western blot and qPCR analysis. Protein levels and a representative western blot of Nrf2 and its downstream related factors, including HO-1 and NQO1 in transfected 3T3-L1 adipocytes (NK, Nrf2- knockdown) were reduced to 21.7 ± 1.6% (P<0.05), 20.1 ± 0.9% (P<0.05) and 29.9 ± 1.9% (P<0.05) (Figures 1C and 1D) and mRNA levels were reduced to 9.3 ± 0.5% (P<0.05) (Figure 1E), 10.3 ± 0.4% (P<0.05) (Figure 1F) and 35.2 ± 1.3% (P<0.05) (Figure 1G), respectively of that of untransfected control cells (Con, control).
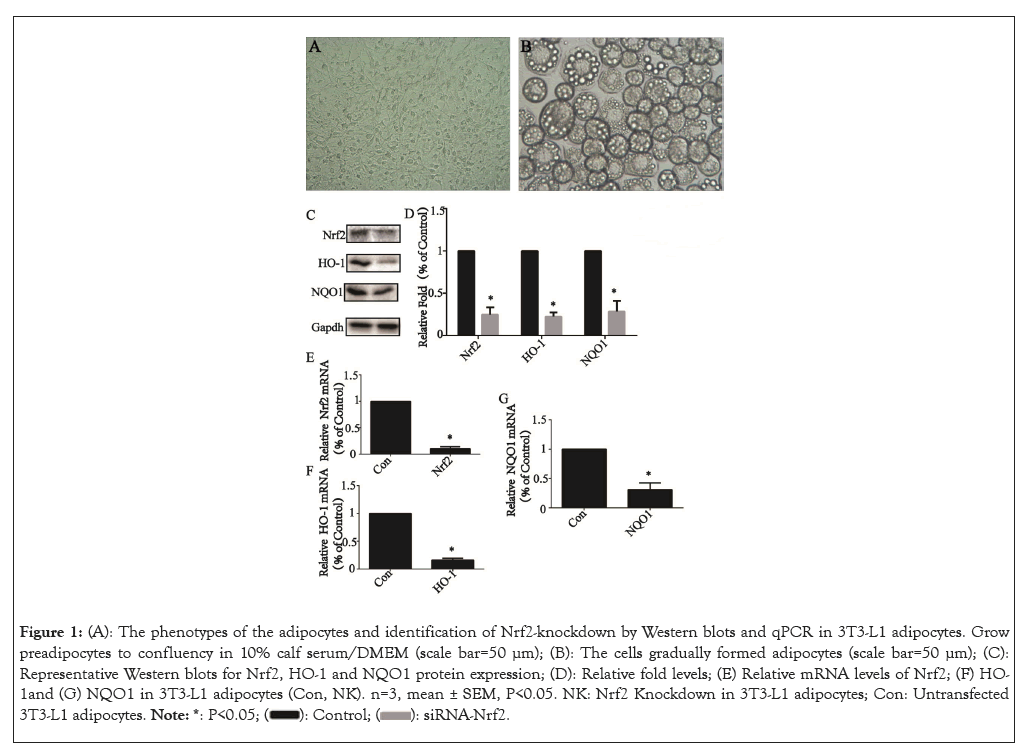
Figure 1: (A): The phenotypes of the adipocytes and identification of Nrf2-knockdown by Western blots and qPCR in 3T3-L1 adipocytes. Grow preadipocytes to confluency in 10% calf serum/DMEM (scale bar=50 μm); (B): The cells gradually formed adipocytes (scale bar=50 μm); (C): Representative Western blots for Nrf2, HO-1 and NQO1 protein expression; (D): Relative fold levels; (E) Relative mRNA levels of Nrf2; (F) HO-1and (G) NQO1 in 3T3-L1 adipocytes (Con, NK). n=3, mean ± SEM, P<0.05. NK: Nrf2 Knockdown in 3T3-L1 adipocytes; Con: Untransfected 3T3-L1 adipocytes. 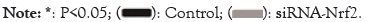
Nrf2 knockdown elevated the expression of BAT marker genes in 3T3-L1 adipocytes
Next, the impact of Nrf2-knockdown on the expression of BAT marker genes were evaluated by examining the protein expression of markers, including PGC-1α, DIO2 and PRDM16 in 3T3- L1 adipocytes. After Nrf2-knockdown, the PGC-1α, Dio2 and PRDM16 protein levels were all significantly elevated (P<0.05) (Figure 2A) and the relative mRNA levels of the genes were also elevated (P<0.05) (Figure 2B), suggesting that Nrf2-knockdown elevated the expression of BAT marker genes in 3T3-L1 adipocytes.
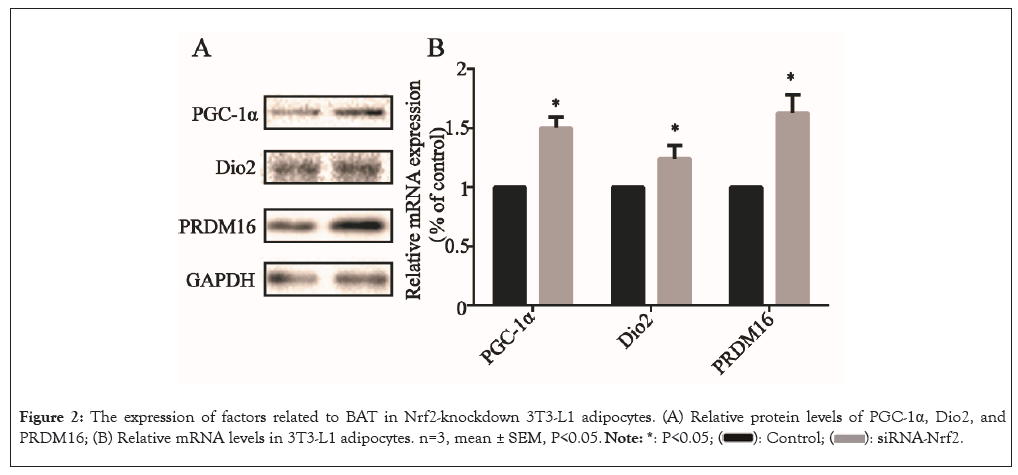
Figure 2: The expression of factors related to BAT in Nrf2-knockdown 3T3-L1 adipocytes. (A) Relative protein levels of PGC-1α, Dio2, and PRDM16; (B) Relative mRNA levels in 3T3-L1 adipocytes. n=3, mean ± SEM, P<0.05. 
Nrf2 knockdown reduced the expression of WAT marker genes of 3T3-L1 adipocytes
We then examined the effects of Nrf2 knockdown on the expression of genes involved in the WAT marker genes, including BMP4, Resistin and Rb1. The relative protein levels of BMP4, Resistin and Rb1 were all lowered upon NK treatment (P<0.05) (Figures 3A and 3B) and the relative mRNA levels of BMP4 (Figure 3C), Resistin (Figure 3D) and Rb1 (Figure 3E) were all lowered upon NK treatment (P<0.05), similar to alterations at the protein level.
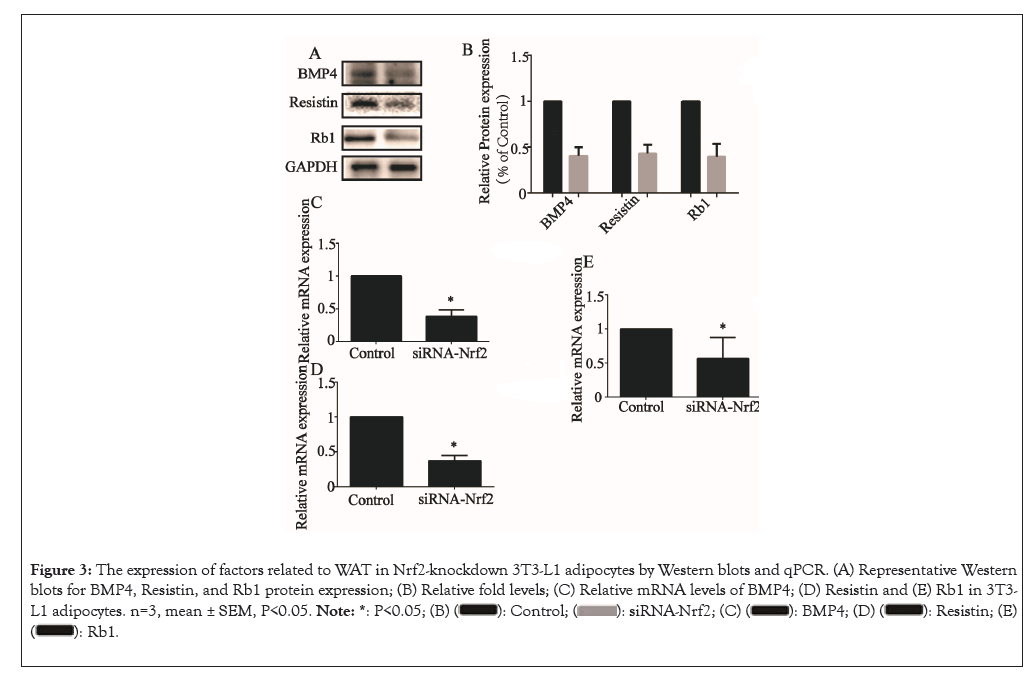
Figure 3: The expression of factors related to WAT in Nrf2-knockdown 3T3-L1 adipocytes by Western blots and qPCR. (A) Representative Western blots for BMP4, Resistin, and Rb1 protein expression; (B) Relative fold levels; (C) Relative mRNA levels of BMP4; (D) Resistin and (E) Rb1 in 3T3-L1 adipocytes. n=3, mean ± SEM, P<0.05. 
Nrf2 knockdown altered the expression of genes which were core functional unit in metabolic control in AT
The impacts of Nrf2-knockdown on the expression of mitochondria related genes, such as Cycs and Uncoupling protein 1 (UCP1), which were the core functional unit in metabolic control in AT. After Nrf2-knockdown, the Cycs and UCP1 protein levels were all significantly elevated (P<0.05) (Figures 4A and 4B) and the relative mRNA levels of the genes Cycs (Figure 4C) and UCP1 (Figure 4D) were also elevated (P<0.05), suggesting that Nrf2-knockdown elevated the expression of regulating metabolic factors of AT in 3T3-L1 adipocytes.
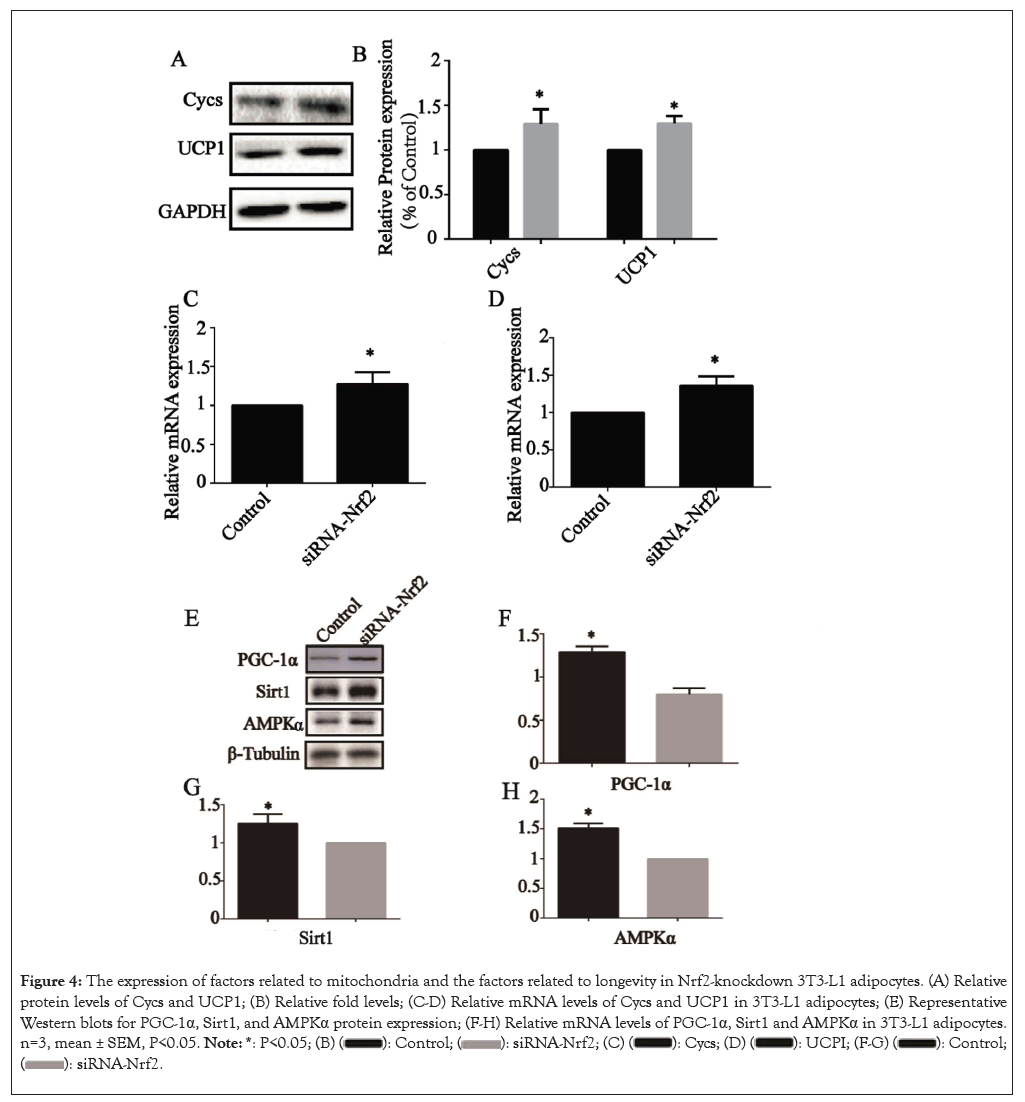
Figure 4: The expression of factors related to mitochondria and the factors related to longevity in Nrf2-knockdown 3T3-L1 adipocytes. (A) Relative protein levels of Cycs and UCP1; (B) Relative fold levels; (C-D) Relative mRNA levels of Cycs and UCP1 in 3T3-L1 adipocytes; (E) Representative Western blots for PGC-1α, Sirt1, and AMPKα protein expression; (F-H) Relative mRNA levels of PGC-1α, Sirt1 and AMPKα in 3T3-L1 adipocytes. n=3, mean ± SEM, P<0.05. 
Nrf2 knockdown increased the expression of proteins involved in longevity gene signaling pathway
The effects of Nrf2-knockdown on the longevity gene signaling pathway was examined in 3T3-L1 adipocytes after Nrf2 transfection. The genes involved in longevity gene signaling pathway, including PGC-1α, Sirt1 and AMPKα. The relative protein levels were all altered upon NK treatment (P<0.05) (Figure 4E) and the relative mRNA levels of PGC-1α (Figure 4F), Sirt1 (Figure 4G) and AMPKα (Figure 4H) were all altered upon NK treatment (P<0.05), similar to alterations at the protein level.
It has long been assumed that the function of adipocytes is simply to store lipids. But now we know that the adipocytes also have important endocrine functions. AT play a role in various physiological and pathological processes such as metabolism and senescence [15]. To better understand the function of adipocytes, it is necessary to study the differentiation of adipocytes. Adipocyte differentiation is a very complex process involving many transcription factors and adipocyte marker genes [16]. A growing body of research has shown that oxidative stress is an important factor in adipogenesis [17,18]. Nrf2 is a key regulatory substance for cells involved in oxidative stress response [19]. Under oxidative stress, Nrf2 dissociates from Keap1 and plays a variety of functions [20]. Several studies have shown that Nrf2 plays an important role in adipocyte generation by regulating the expression of various factors [21]. Different from white adipocytes, brown adipocytes have high expression of DIO2, PRDM16 and PGC1α. PRDM16, a transcription co-regulator responsible for brown adipocyte development in classic BAT depot, has unique roles in browning capacity [22]. Where its overexpression markedly promoted browning in a cell-autonomous manner likely through de novo adipogenesis [23]. Also, the activation of PGC-1α has a similar effect [22]. Our results showed that expression of BAT marker genes increased after silencing Nrf2 in 3T3-L1 adipocytes. Therefore, we speculate that it may improve cell senescence and other processes based on it.
Mitochondria are the core functional unit in metabolic control in numerous cells, including BAT and WAT [24]. In the context of aging, it has been reported that mitochondrial enzyme expression is reduced in adipose tissue from old mice, yet little is known regarding mechanisms that could be mediating these changes [25]. Similarly, it is well established that human WAT mitochondrial function measured by oxygen consumption of the tissue is reduced both in obesity as well as during aging [26]. In addition, mitochondria have been suggested as key components in the process of white adipocyte formation through Reactive Oxygen Species (ROS) signaling [27]. Furthermore, mitochondrial function has been linked to the white adipocyte endocrine functionality as it was shown that secretion of adipokines such as adiponectin are dependent on mitochondria [28]. BMP4, Resistin and Rb1 are marker genes of WAT [29]. When Nrf2 is knocked down or knocked out, production of WAT is significantly reduced, but a series of more severe metabolic syndromes are induced, including insulin resistance, hyperlipidemia and rapid glucose absorption [30]. Nrf2 controls formation of WAT, insulin sensitivity and glucose and lipid homeostasis by regulating the generation and function of fat [31]. NAD(P)H quinone oxidoreductase 1 (NQO1) and Heme Oxygenase-1 (HO-1) are key target genes of Nrf2 downstream against oxidative stress [32]. In addition, important studies have also pointed out that reducing Visceral Adipose Tissue (VAT) content can resist the occurrence of age-related diseases through BAT activation [33]. Our results were also consistent with previous studies, indicating that marker genes of WAT were decreased after knockdown of Nrf2, reflecting the regulatory effect of Nrf2 on WAT generation. These results also further confirmed the role of Nrf2 on adipose transformation and anti-aging.
Mitochondrial electron transport is the main source of ROS, making the mitochondrial network the most immediate target of oxidative damage that can lead to mitochondrial dysfunction.
On the other hand, several studies clearly refute the current model that ROS may be involved in the senescence-related decline of mitochondrial function [34]. In summary, a decline in mitochondrial energy metabolism and the accumulation of mitochondrial DNA (mtDNA) mutations are important contributors to human aging. This decline in mitochondrial function has also been reported in BAT and is thus congruent with similar observations in other tissues [35]. Studies have shown that, it is tempting to speculate that the induction of a brown fat phenotype, which would result in an increased mitochondrial functionality, could be the reason for the observed beneficial effects on aging and metabolism [36]. Our results also showed that the expression of mitochondria related genes increased after Nrf2 knockdown, which may play a possible role in improving the aging process.
AT also exerts global effects on inflammatory signaling and insulin sensitivity through the secretion of adipokines and adipogenic hormones [37]. Moreover, adipose tissue-specific dysfunctions with impact on the development of metabolic syndrome and deleterious effects on whole-body energy homeostasis are also related to aging [38]. Most importantly, key physiological processes AT affect molecular pathways that regulate lifespan. For example, levels of Sirt1 decline with age in several tissues, including AT and this decline is exacerbated in mouse models of accelerated aging leading to senescent obesity, impaired Browning and metabolic syndrome. UCP1 also has a similar physiological and pathological process [14]. In our study, the expression of these anti-aging factors was increased in 3T3-L1 adipocytes by Nrf2 knockdown, which may also play an important role in delaying the process of aging.
Taken together, many of the classic mechanisms of aging, such as cellular senescence, chronic inflammation and mitochondrial dysfunction, occur in AT. Thus, emerging interventions targeting aging mechanisms such as oxidative stress should have an impact on AT. We investigated that Nrf2 a key regulator of oxidative stress, might play an anti-aging role through this form, but further studies are needed. This opens up the possibility of using AT as an indicator of the efficacy of such therapies. In addition, therapies that extend life and target the basic mechanisms of aging often have a significant impact on AT. Conversely, it may also be possible to use our knowledge about AT aging to design therapies that specifically target the mechanisms of adipose aging. Because AT function is closely related to insulin sensitivity and inflammation, targeting AT aging may be a new frontier in the treatment of age-related type 2 diabetes, metabolic syndrome and its complications. Thus, AT may be a powerful therapeutic target for the development of new therapies against aging and aging- related diseases.
This study aimed at whether miR-365 could regulate ferroptosis of gastric cancer cells. Results showed that miR-365 downregulated in cell lines and tissues. Moreover, expression of miR-365 mimics enhanced the erastin-induced ferroptosis in gastric cancer cells. Additionally, miR-365 mimics further upregulated the MDA, Fe2+ , ROS levels in SGC-7901 and MGC- 803 cells after erastin exposure and antioxidative GSH levels were further downregulated, which indicate that lipid oxidation in gastric cancer cells induced by erastin could be enhanced by overexpressing miR-365. Nrf2 took part in the ferroptosis of cancer cells, it was observed in gastric cancer cells after erastin exposures. qRT-PCR and Western blot results showed that erastin increased the transcription and translation levels of Nrf2 in gastric cancer cells. The luciferase data confirmed that miR- 365 directly targets Nrf2. To compare with NC simulation, miR- 365 simulation could reduce the activity of luciferase to Nrf2 3’- UTR wild type but no effect on the mutation type, which could provide evidence that miR-365 could target Nrf2.
This research was supported by the Youth Natural Science Foundation of Hebei Province (grant no: H2020206163); The key Scientific and technological research program of Hebei Health Commission (grant no: 20200122).
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Yuan X, Wang H, Xie B, Ding Y (2024) Nuclear Factor E2-Related Factor 2 Knockdown Alters Adipose-Phenotype and Increases Expression of Genes Involved in Longevity in 3T3-L1 Cells. J Clin Chem Lab Med.7:285.
Received: 02-Nov-2023, Manuscript No. JCCLM-23-27884; Editor assigned: 06-Nov-2023, Pre QC No. JCCLM-23-27884; Reviewed: 20-Nov-2023, QC No. JCCLM-23-27884; Revised: 27-Nov-2023, Manuscript No. JCCLM-23-27884; Published: 04-Jan-2024 , DOI: 10.35248/2736-6588.24.7.285
Copyright: © 2024 Yuan X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.