
Journal of Clinical Toxicology
Open Access
ISSN: 2161-0495

ISSN: 2161-0495
Research Article - (2023)Volume 13, Issue 2
siRNA is a promising tool for targeted therapy. Previously we conjugated the siRNA to Squalene (SQ) by copper- free click chemistry, which has been proven to successfully deliver siRNA to tumors with fusion oncogenes and Schwann cells. To go further for future human application, this study aims to optimize the synthesis and the nanoprecipitation of siRNA-SQ conjugates while conserving their physicochemical characteristics and efficiency. We succeeded in increasing the yield of the siRNA-SQ conjugation reaction making it scalable up to 50-times. The size, the polydispersity index and the ζ-potential of the nanoparticles obtained are strongly dependent on the solvent used. Moreover, the molecular dynamics simulations reveal the physical mechanisms of nanoparticle formation at the early stage of their growth. However, these parameters do not influence the activity of the nanoparticles both in vitro and in vivo. Established protocols of efficient and reliable siRNA-SQ nanoparticles production open up the perspective of their therapeutic usage in humans.
Squalenoyl siRNA nanoparticles; Physicochemical properties; Molecular simulation; In vitro and in vivo studies
The RNA interference discovery by Fire A, et al. [1], in 1998 transformed the therapeutic world. They depicted that small double-stranded RNAs (siRNA), were able to decrease the mRNA expression of a targeted gene. siRNAs are small double- stranded RNAs of 19-21 base pairs with 3’-cohesive ends and a molecular weight of 13 kDa [2]. They inhibit gene expression and subsequently, protein translation by activating the RNA-Induced Silencing Complex (RISC). Through the action of the catalyst Ago2 the targeted mRNA sequence is cleaved [3]. siRNA is specific and active at low doses making them an attractive tool to develop new therapeutic approaches. However, there are major obstacles to their use: 1) Delivery to targeted cells or organs due to their physicochemical characteristics (hydrophilic and degradation by nucleases), 2) Dose-related toxicity and 3) Off-target effects [4]. These challenges can be addressed by the development of nanocarriers such as cationic lipids and polymers, or conjugation to a lipophilic molecule made it possible to counteract these hurdles. The role of such nanoobjects is protecting siRNA from degradation, promoting cellular uptake, and avoiding the innate immune response [4,5]. In 2018, the first siRNA drug-related content was approved by FDA for the treatment of hereditary transthyretin-mediated amyloidosis [6]. Later on, in 2020, givosiran (commercial name givlaari) was accepted as a treatment for acute hepatic porphyria [7]. Despite these successful cases of siRNA delivery to the hepatic tissue, there is still a great demand for safe and efficient siRNA nanocarriers for other tissues and cell types.
We have previously developed the squalenoylation method which is based on the conjugation of siRNA to a biodegradable Squalene (SQ) molecule [8], a natural precursor of cholesterol. Such conjugates form Nanoparticles (NPs) in an aqueous solution spontaneously [9]. Initially siRNA was coupled to SQ using maleimide-sulfhydryl chemistry and the resulting NPs showed significant activity in xenograft mice models of papillary thyroid carcinoma (RET/ PTC1 and RET/PTC3 oncogenes) or prostate cancer (TMPRSS2- ERG oncogene). However, the yield of this conjugation reaction was very low [8,10-12], which limited its applicability. Later, the method of conjugation was improved using the copper-free click chemistry to conjugate the siRNA TMPRSS2-ERG to SQ [13]. This bio-orthogonal reaction increased the yield of conjugation and preserved the activity of the synthesized siRNA TMPRSS2-ERGSQ NPs in the xenografted prostate cancer mice model. Recently, we succeeded to deliver the siRNA PMP22-SQ NPs that targeted the PMP22 overexpression in Charcot Marie tooth 1A disease, one of the most common forms of inherited peripheral neuropathies [14]. Further studies on the fate of the siRNA-SQ NPs in blood showed that they interact with serum albumin and low-density lipoproteins that facilitate the transport of siRNA-SQ NPs to the target tissues [15]. These success stories of using siRNA-SQ NPs in animal models stimulated their transfer to humans.
This study aims to improve the siRNA-SQ synthesis and to find the best nanoprecipitation method. The scaling-up of the process was carefully monitored to rule out any potential incomplete synthesis reaction and to identify the best nanoprecipitation method that complies with the standards that we have defined (size<300 nm, polydispersity index<0.3, ζ-potential ≤ -50 mv). Two nanoprecipitation approaches were developed in conjunction with in silico Molecular Dynamics (MD) simulations. Their in vitro and in vivo efficiency was also investigated.
Chemical experiments
siRNA and chemical modifications: The siRNA PMP22 [14], and the siRNA Ctrl (scrambled sequence) used in this study were purchased as single-stranded RNA from Eurogentec France. To provide stability to the single-stranded RNA they were synthesized as 19-mers with two 3’ overhanging 2’-deoxynucleotide residues as described by Tuschl [16]. To allow the conjugation of the siRNA to SQ by Copper (Cu)-free click chemistry, the sense strand of the siRNAs was modified by adding a Dibenzocyclooctyne (DBCO) reactive group bound through a N-(hexamethylenyl)-6- oxohexanamide (C6) linker at the 5’-end [13].
siRNA-SQ conjugation scale-up: Initially, to conjugate the siRNA to SQ by Cu-free click chemistry, the C6-sense siRNA strand was added to N3-SQ at a ratio of 1:50 in a solution of acetone and DMSO and then incubated for 12 h under stirring. After that excess acetone was removed under nitrogen flux [13]. Then the conjugate siRNA-SQ was purified by reverse-phase HPLC to remove the excess of N3-SQ. To scale up the siRNA-SQ conjugation step, the N3-SQ quantity used was first optimized by screening the ratios of the C6-sense siRNA strand to N3-SQ. To this aim, the 1:50, 1:25, 1:12.5, and 1:5 ratios were tested. Having identified the optimal ratio (sense siRNA/N3-SQ: 1:25) the second step of scale-up to increase the quantity of sense siRNA PMP22 was initiated. The increasing amount of the C6 sense siRNA PMP22 from 10 nmol, to 20 nmol, 100 nmol and 500 nmol were evaluated still using the 1:25 sense siRNA/N3-SQ ratio. The following steps of the original preparation were not changed.
siRNA nanoparticle preparation: To prepare the double-stranded siRNA-SQ, equimolar amounts of sense-siRNA PMP22-SQ or siRNA Ctrl-SQ and antisense strands (siPMP22 or siCtrl) were added to a hybridization buffer (30 mM HEPES-KOH (pH 7.4), 2 mM Mg acetate, 100 mM K acetate). The solution was incubated for 3 min at 95°C, then for 45 min at Room Temperature (RT).
The nanoprecipitation was performed in two different methods. The first method called “forward” was previously published by our team [13-15]. Briefly, the annealed siRNA-SQ bioconjugates in UltraPureTM distilled water (DNase- and RNase-free water) were added dropwise to an organic phase (acetone) under stirring with a volume/volume ratio of 1 (organic phase): 2 (aqueous phase). Then acetone was evaporated using nitrogen flux for 30 min to obtain an aqueous suspension of pure siRNA-SQ NPs. The second method of nanoprecipitation is called “inverse” in which the organic phase (acetone) was added drop by drop over stirring to the siRNA-SQ aqueous solution with a volume/volume ratio of 1 (organic phase): 2 (aqueous phase). Then the acetone was removed as previously under 30 min nitrogen flux. To increase the concentration of the siRNA-SQ NPs from 10 μM to 50 μM, 50 nmol of C6-sense siRNA PMP22-SQ were annealed to 50 nmol of antisense siRNA PMP22 as described above then nanoprecipitated using the “inverse” method.
siRNA SQ physicochemical characteristics and stability measurements: NPs size (hydrodynamic diameter), Polydispersity Index (PDI) and surface charges (-potential) were measured by Dynamic Light Scattering (DLS) using Malvern Zetasizer Nano ZSP (Malvern Instrument) with a scattering angle of 173°. To study the stability at 4 °C and at RT, hydrodynamic diameter, PDI and -potential of the siRNA-SQ NPs were measured at different time points.
Modeling of siRNA-SQ NPs
The Coarse Graining (CG) procedure: MARTINI force field version 2.1 adapted for nucleic acids [17], was used. The coarsegraining of SQ and the linker is challenging because their molecular structures are incompatible with the usual 4-1 mapping used for the Martini force field. We used an approach with overlapping CG beads of different sizes as described in Supplementary Information. The parameterization of obtained CG topology follows the recommended procedure of mapping CG bonds and angles to the corresponding values computed for the reference all-atom trajectories (see Supplementary Images (SI) for details, Supplementary Figures S1-S5).
Molecular Dynamics (MD) simulations: The all-atom topology of the spacer was generated with the CHARMM GUI ligand builder [18,19]. The all-atom topology of SQ was used from previous work [20,21]. CHARMM36 force field [22], was used for all components of the all-atom system. Martini v2.1-dna force field (http:// cgmartini.nl/index.php/rna) [18], was used for all CG simulations. All simulations were performed in Gromacs [23], version 2019.2. The all-atom and CG simulation protocols are detailed in SI, Details of MD simulations.
RNA modeling: The fd_helix program (https://casegroup.rutgers. edu/fd_helix.c) [24] was used to build the ideal A-form doublestranded RNA from the AAAUACCAACUGUGU GGACUATT sequence. The trailing TT nucleotides were left dangling in both strands. The atomistic structure was converted to CG representation by the martinize-nucleotide.py script (http://cgmartini.nl/ index.php/rna). The RNA, spacer and squalene topologies were combined as described in SI, Combining topologies paragraph.
Simulations of the conjugate self-assembling: Either 30 or 100 CG conjugates were randomly placed into the empty cubic box with dimensions 25 × 25 × 25 nm. The remaining space was filled with the solvent beads and the corresponding number of randomly placed Na+ counter ions. Either pure water or pure acetone solvents were used. This resulted in 4 production systems encoded as 30 w, 30 a, 100 w, and 100 a where the number stands for the number of conjugates, and the letters represent the solvent (“w” for water, “a” for acetone). Each system was energy minimized and simulated for 6-10μs depending on the equilibration rate. The analysis was performed by the custom plugin for Pteros molecular modeling library [25]. The centers of masses of squalene moieties of all conjugates were subject to agglomerative clustering with the linkage by minimal distance and the distance cut-off of 2 nm. The number of clusters and their average size were monitored at each trajectory frame.
Biological experiment
Cell line: MSC80 cell line (mouse Schwann cell line) that expresses myelin genes PMP22 and P0 was used in this study [26]. Cells were grown in a complete DMEM medium supplemented with 10% heat-inactivated FBS, with 100 units/mL penicillin, 100 μg/mL streptomycin and 2.5 μg/ml of Fungizone Amphotericin. Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
In vitro cell transfection: 3 × 105 MSC80 cells were seeded in six-well plates containing complete medium until 60%-70% confluency. Then, transfection was carried out using Lipofectamine 2000® according to the manufacturer’s instructions in Opti-MEM reduced serum-free medium. Naked siRNA, siRNA Ctrl-SQ NPs “forward” and siRNA Ctrl-SQ NPs “inverse”, siRNA PMP22-SQ NPs “forward” and siRNA PMP22-SQ NPs “inverse” were transfected at 50 nM concentration. Four hours later, the medium was replaced with a complete DMEM medium. After 48 h, cells were harvested, then mRNA was extracted to determine gene expression.
mRNA extraction and Real-Time PCR (RT-qPCR): Using an RNeasy mini-kit (Qiagen, Courtaboeuf, France) total RNA was extracted from MSC80 cells. Reverse transcription was performed by M-MLV RT buffer pack (Invitrogen, Charbonnières-les-Bains, France). Real-Time quantitative PCR (RTqPCR) was carried out with the CFX96TM Real-time system (Biorad) using “Maxima Syber Green Rox qPCR master Mix” (Thermo Scientific, Villebonsur- Yvette, France), according to the manufacturer’s instructions. PMP22 gene expression was determined by the 2−ΔΔCt method and normalized to a control gene level 18 S. Relative mRNA expression of the targeted gene was compared to non-treated cells. The experiment was repeated at least 2 times.
Transgenic mouse model and animal protocol: The C61 Charcot Marie Tooth 1A (CMT1A) transgenic mouse model which carries 4 extra copies of the human PMP22 gene and shows mild demyelination with an intermediate nerve conduction velocity was used in this study [27].
At the age of 8 weeks C61 mice were divided into three groups of three mice each. Group one received 5% dextrose solution as a vehicle, mice of group two were treated with siRNA PMP22-SQ NPs prepared with the “forward” nanoprecipitation method, and mice of the third group were treated with siRNA PMP22-SQ NPs “inverse” nanoprecipitation method. In addition, one group of WT B6 mice was used as a control (4 mice). All the treatments were administered by i.v. the route, following a 3-injection treatment schedule of 0.5 mg/kg per injection. Each injection was separated by three days. Behavioral studies and mearsurements of the Compound Muscle Action Potential (CMAP) were done before the start of the treatment and at the end.
The number of animals per group, is in accordance with the 3R rule that aims to reduce the use of animals in preclinical research. All animal experiments were approved by the institutional Ethics Committee of Animal Experimentation and research council, registered in the French Ministry of Higher Education and Research «Ministère de l’Enseignement Supérieur et de la Recherche; MESR, autorisation N°: APAFIS#10131-2016112916404689». It is carried out according to French laws and regulations under the conditions established by the European Community (Directive 2010/63/ UE). The investigation has been conducted in accordance with ethical standards and according to the Declaration of Helsinki. All efforts were made to minimize animal suffering. Administration of treatments was performed under isoflurane anesthesia. All animals were housed in a sterilized laminar flow caging system. Food, water, and bedding were sterilized before animals were placed in cages. Food and water were given ad libitum.
Behavioral tests
Locotronic test: To test the locomotion of the C61 mice before and after treatment, a locotronic apparatus (Intellibio innovations A-1805-00049) was used. The test was performed as previously described [14], where the time taken to perform the test by each mouse was recorded. The test was repeated three times for each mouse and the average time was recorded. Data are presented as the mean ± S.D.
Grip strength test: To assess the muscular activity of the C61 mice before and after treatment, an automated grip strength meter (BIOSEB Innovation Model: BIO-GS3) was used as previously described [14]. The maximum force in grams of the forelimbs and total limbs was recorded for each mouse. Data are presented as the mean ± S.D.
Electrophysiological test Compound Muscle Action Potential (CMAP): The test was performed as previously described [14], with a standard EMG apparatus (Natus UltraPro S100 EMG) in accordance with the guidelines of the American Association of Neuromuscular and Electrodiagnostic Medicine. Briefly, for recording the CMAP level, the mice were placed on their frontal side on a heating pad to maintain their body temperature between 34°C and 36 °C and kept under anesthesia. The stimulator needle electrode was placed at the sciatic nerve notch level, the anode electrode in the upper base part of the tail, while the receptor needle (or recording needle) was inserted in the medial part of the gastrocnemius muscle. Then a supramaximal square wave pulse of 8 mA was delivered through the stimulator needle and recorded by the receptor needle through the muscle as an amplitude. Data are recorded as mean ± S.D.
Statistical analysis
Statistical analysis was computed using GraphPad Prism 8.3.0 software. Mann-Whitney analysis was performed to assess the statistical difference between two groups. For grouped analyses, a one-way ANOVA followed by Bonferroni or Kruskal Wallis multiple comparison test was performed. A value of p<0.05 was considered significant.
Squalene concentration plays a key role in the conjugation of C6-sense strand siRNA PMP22 to the squalene molecule
In all our previously published data the conjugation step was performed using 10 nmol C6-sense strand siRNA PMP22 and 500 nmol of the N3-SQ solution prepared in DMSO. The C6 sense siRNA PMP22-SQ conjugate was purified by RP-HPLC to remove the excess of N3-SQ. The siRNA sense-SQ peak was obtained at a retention time of 16 min as shown in the chromatogram (Supplementary Figure S6A) [14,15]. Using the same molar ratio of 1: 50 of sense siRNA: N3-SQ we tried to increase the number of moles of the sense siRNA to be conjugated to SQ up to 100 times. However, such a scaling up induced the formation of polar by-products as evidenced by several peaks on the HPLC graph that appeared between 13 and 15 min in addition to the purified sense siRNA-SQ at 16 min (Supplementary Figure S6B). This caused a drastic 40% reduction in the yield of the siRNA-SQ sense strand.
Hence, to improve the yield of the chemical reaction and to be able to increase the concentration of the bioconjugate siRNA sense-SQ we had to adjust the quantities of the sense siRNA and N3-SQ in the conjugation step. Therefore, different ratios of sense siRNA PMP22 to N3-SQ were tested and summarized as follows: 1 nmol C6-sense siRNA PMP22: 5 nmol of N3-SQ, 1 nmol C6-sense siRNA PMP22: 12.5 nmol of N3-SQ, 1 nmol C6-sense siRNA PMP22: 25 nmol of N3-SQ compared to the original ratio which was 1 nmol C6-sense siRNA PMP22: 50 nmol of N3-SQ. Our results showed that the best ratio of sense siRNA PMP22: N3-SQ is 1:25 as it gave higher peaks on the HPLC at an elution time of 16 min and a higher yield of 90% (Supplementary Figure S6C). Using this new ratio between N3-SQ and the sense siRNA PMP22 strand we were successful in increasing the amount of the sense strand conjugated to N3-SQ from 10 nmol to 20 nmol (Supplementary Figure S6D), 100 nmol (Supplementary Figure S6E), and even 500 nmol (Supplementary Figure S6F). All the HPLC graphs showed a single peak at the elution time of 16 min which confirmed the purity of the bioconjugate despite having a small peak at 10 min which is the unconjugated sense strand. The reaction yield after HPLC purification is approximately 90%.
The method of nanoprecipitation influences the size of the nanoparticles
In all our previous work the siRNA-SQ nanoparticles were obtained by a “forward” nanoprecipitation method. The aqueous solution containing the siRNA-SQ bioconjugates was added to an organic solvent (acetone). This procedure was performed at a concentration of 10 μM and resulted in NPs of an average hydrodynamic diameter ranging between 180 nm to 280 nm with a PDI<0.3 and a -potential ≤ -40 (Figures 1A-1C black square). To assess whether we can decrease the size of the NPs, we changed the nanoprecipitation method in which the organic solvent was added to the aqueous siRNA-SQ solution. This “inverse” nanoprecipitation method leads, at the same concentration (10 μM), to the formation of NPs of 80 nm diameter, PDI<0.14 and a -potential of<-35 (Figures 1A-C white square). The cryo-TEM images confirmed DLS measurements showing spherical compact nanoobjects of 167 ± 31 nm for the “forward” nanoprecipitation (Figure 1D) and 95 ± 14 nm for the “inverse” nanoprecipitation (Figure 1E). This confirmed that the “inverse” method of the nanoprecipitation allowed decreasing hydrodynamic diameter without statistically affecting -potential.
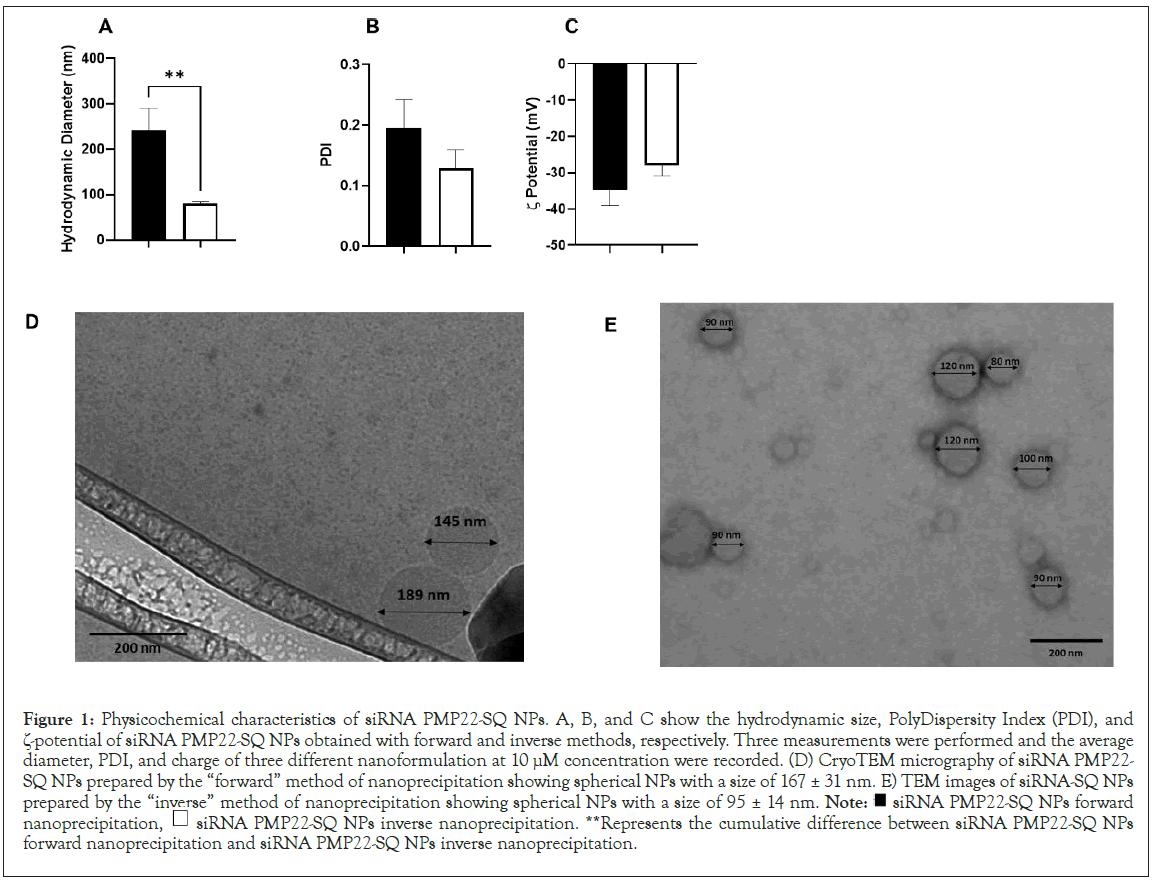
Figure 1: Physicochemical characteristics of siRNA PMP22-SQ NPs. A, B, and C show the hydrodynamic size, PolyDispersity Index (PDI), and
ζ-potential of siRNA PMP22-SQ NPs obtained with forward and inverse methods, respectively. Three measurements were performed and the average
diameter, PDI, and charge of three different nanoformulation at 10 μM concentration were recorded. (D) CryoTEM micrography of siRNA PMP22-
SQ NPs prepared by the “forward” method of nanoprecipitation showing spherical NPs with a size of 167 ± 31 nm. E) TEM images of siRNA-SQ NPs
prepared by the “inverse” method of nanoprecipitation showing spherical NPs with a size of 95 ± 14 nm. Note:  siRNA PMP22-SQ NPs forward
nanoprecipitation,
siRNA PMP22-SQ NPs forward
nanoprecipitation,  siRNA PMP22-SQ NPs inverse nanoprecipitation. **Represents the cumulative difference between siRNA PMP22-SQ NPs
forward nanoprecipitation and siRNA PMP22-SQ NPs inverse nanoprecipitation.
siRNA PMP22-SQ NPs inverse nanoprecipitation. **Represents the cumulative difference between siRNA PMP22-SQ NPs
forward nanoprecipitation and siRNA PMP22-SQ NPs inverse nanoprecipitation.
The scale-up did not affect the hydrodynamic diameter and the PDI of nanoparticles
Our results suggest that the scale-up of the sense-siRNA PMP22-SQ synthesis at 50 μM followed by hybridization with the antisense strand and “inverse” nanoprecipitation did not affect both the hydrodynamic diameter and the PDI of the nanoparticles. Thus, with this new protocol we were able to produce at 50 μM, siRNA PMP22-SQ NPs of 154.2 ± 0.8 nm size with a low PDI of 0.13 ± 0.01.
NPs produced by both methods are stable at room temperature
The stability of nanoparticles is an important criterion that needs to be taken into consideration in practical applications. To assess the stability, the hydrodynamic diameter and the PDI of the siRNASQ NPs were measured by the DLS at various time points when stored at Room Temperature (RT). The size, PDI, and -potential were stable for the nanoparticles produced using both the forward (Figure 2A and 2B) and the inverse (Figure 2C and 2D) methods. However, we noticed that after 13 days at room temperature, mold was found in the solution.
Figure 2: Stability of siRNA-SQ NPs at room temperature. (A and C) Kinetic of hydrodynamic diameter and PDI of siRNA-SQ NP using “forward”
(A) or “inverse” (C) methods of nanoprecipitation. (B and D) Kinetic of ζ-potential of siRNA-SQ NPs synthesized by the “forward” method (B) or
“inverse” method (D) of nanoprecipitation. Each point represents the average of 3 different nanoformulations ± SD. No statistical significance was
found for size, PDI, and ζ-potential. Note:  Hydrodynamic diameter,
Hydrodynamic diameter,  PDI,
PDI,  ζ-potential.
ζ-potential.
At 4°C both formulations are still injectable after one year
At 4°C, the size of the siRNA PMP22-SQ NPs prepared using the forward method showed only a slight increase in size after 12 months without any change in the PDI (Table 1). In contrast, the siRNA PMP22-SQ NPs prepared by the inverse method, showed a significant decrease in the size and the PDI after one year (Table 1). Despite this significant change in the size of NPs, both formulations are still in the injectable size range after one year of storage.
| Diameter (nm) | PDI | |||||
|---|---|---|---|---|---|---|
| Time | 0 month | 1 month | 12 months | 0 month | 1 month | 12 months |
| siRNA-SQ NPs "forward" | 236.9 ± 29,1 | 271.2 ± 64.1 | 276.8 ± 16* | 0.26 ± 0.07 | 0.27 ± 0.1 | 0.24 ± 0.06 |
| siRNA-SQ NPs "inverse" | 102.9 ±16 | 115.46 ± 18.4 | 82.71 ± 0.9**$$$ | 0.19 ± 0.06 | 0.22 ± 0.02 | 0.15 ± 0.04$$ |
Note: * Statistical difference between zero months and 12 months; $ The statistical difference between one month and 12 months. Data represents Mean ± SD. *p<0.05; **, $$ p<0.01, ***, $$$ p<0.001 using one-way ANOVA followed by Kruskal Wallis multiple comparison test.
Table 1: Stability at 4°C of siRNA-SQ NPs prepared using both methods of nanoprecipitation.
The structure of the resulting siRNA PMP22-SQ NPs depend on the nanoprecipitation solvent
In the forward method, the formation of nanoparticles starts in acetone while in the inverse one it starts in almost pure water. Although MD simulations do not replicate the nanoprecipitation protocol exactly due to time and scale limitations, they are able to provide useful insights into the very early first stages of growth in the corresponding water or acetone environments. All MD simulations showed a clear tendency of aggregation both in acetone (forward) and water (inverse) nanoprecipitation methods. However, the structures of resulting complexes (and hence their size) depend strongly on the solvent. The aggregation always occurs by squalene moieties, which stick together and form a micelle with a compact hydrophobic core surrounded by a loose “crown” of RNA helices. However, the number of molecules in such micelle and their arrangement differ substantially in water and acetone (Figure 3).
Figure 3: The structures of typical individual micellar clusters of conjugates formed in water (A) and acetone (B). Each conjugate molecule is shown in a different color. The CG beads of squalene moieties are shown as spheres. The RNA moieties are shown as the networks of CG bonds with semitransparent molecular surfaces. The clusters form larger aggregates facilitated by RNA interactions in water (C) and in acetone (D). Panels C and D show the whole simulation box with periodic boundary conditions.
In water i.e., inverse nanoprecipitation stable dimers and trimers of conjugates are formed, while larger clusters are unstable and only form transiently (Figure 3A). Although the hydrophobic squalene core is still exposed to water in such arrangement, no further merging of dimers and trimers occurs on the time scale of ~10 μs. In contrast, in acetone i.e. forward nanoprecipitation much larger stable micellar clusters, containing up to 8 conjugates, are formed (Figure 3B).
The dimers and trimers in water do not interact with each other and only collide transiently in the course of random diffusion (Figure 3C). In contrast, in acetone individual nano assemblies combine further by means of their RNA moieties and form a loose network, which spans the whole simulation box (Figure 3D). By the end of the simulation, all nanoassemblies are combined in acetone and no freely diffusing aggregates or monomers remain. In acetone formation of the linear rod-like head-to-head, pairs of RNA helices are observed (visible as two-colored linear RNA rods in Figure 3D). The RNA helices in such pairs belong to the same particle and may further form lateral contact with the other nano assemblies.
The size and time for the production of nano assemblies of siRNA PMP22-SQ NPs differ depending on the method of nanoprecipitation
The average size of the micellar clusters increases with simulation time and shows clear signs of saturation (Figure 4). The abrupt jumps on the plots correspond to the rare events of merging or dissociation of clusters in larger aggregates.
Figure 4: Evolution of the mean size of micellar clusters of conjugates in MD simulations. The systems are encoded as 30 w, 30 a, 100 w, and 100 a
where the number stands for the number of conjugates, and the letters represent the solvent (“w” for water, “a” for acetone). Note:  30 w, (
30 w, ( ) 30 a,
) 30 a,  100 w,
100 w,  100 a.
100 a.
The average size of the nano assemblies is about 2 nm in water and 4-6 nm in acetone, which agrees with the visual inspection of trajectories (Figure 3C and 3D). The nano assemblies are forming faster at a higher initial concentration of conjugates (the curves for 100 a, and 100 w are growing faster than the corresponding curves for 30 a, and 30 w), but their final sizes are similar on the available simulation time scale. All curves in Figure 4 show a clear tendency of saturation after ~2 μs (the log scale makes this less obvious but emphasizes the differences in initial simulation stages).
Spacer arrangements differ in the two methods of nanoprecipitation
From the visual inspection, it is evident, that in water the spacer is always folded onto the surface of RNA and sticks to it strongly. As a result, the squalene is attached almost directly to the RNA surface and has very little spatial mobility. In contrast, in acetone the spacer sticks out from RNA and forms a long stem-like linkage between the squalene core of the micelle and the RNA helices. In order to quantify this difference, we computed the number of RNA beads per conjugate, which are in close vicinity (0.6 nm) of squalene moiety (Supplementary Figure S7).
Squalene is in close contact with RNA in water (~16.7 contacts per conjugate), while there are almost no such contacts in acetone (~3.4 contacts per conjugate). This quantity is independent of the number of conjugates in the simulation box (Supplementary Figure S7) and doesn’t change significantly during simulations (data not shown).
Both “forward” and “inverse” nanoprecipitation methods gave active nanoparticles that ameliorate the behavioral and physiological activity of CMT1A mice
Our recent findings showed that treatment using siRNA PMP22- SQ NPs could lead to targeted therapy for CMT1A neuropathy that is caused by a duplication of the PMP22 gene [14]. Based on these findings, we chose the siRNA PMP22 to compare the two nanoprecipitation methods, both in vitro and in vivo. To be sure that the siRNA PMP22 is still active after nanoprecipitation, we first assessed its ability to decrease PMP22 expression in vitro. Therefore, MSC 80 cells that express the PMP22 gene were transfected for 48h by naked siRNA PMP22, “forward” siRNA PMP22-SQ NPs, and “inverse” siRNA PMP22-SQ NPs. The scramble siRNA Ctrl-SQ sequence was transfected similarly using nanoparticles prepared by either the “forward” or the “inverse” nanoprecipitation method. Results have shown that the method of nanoprecipitation does not affect the activity of the siRNA PMP22-SQ NPs reflected by the 50% inhibition of the PMP22 mRNA expression in MSC80 cells. The siRNA Ctrl-SQ NPs did not differ from the non-treated cells (Supplementary Figure S8).
Afterward, we assessed the ability of both formulations on agematched C61 CMT1A mice. Mice were treated via intravenous injection with siRNA PMP22-SQ NPs prepared using both methods of nanoprecipitation in addition to a group that received a vehicle (5% dextrose) and compared to wild-type mice. After treatment, the mice treated with both siRNA PMP22-SQ NPs showed amelioration in the motor activity reflected by the significant decrease in the time taken by the mice to perform the locotronic test, which is comparable to the wild-type mice (Figure 5A). For the muscular activity, both treated groups showed significant improvement as the mice became stronger on both fore and hind limbs compared to the group that just received the vehicle (5% dextrose) (Figures 5B and 5C). Moreover, the electrophysiological activity, tested by the Compound Muscle Action Potential (CMAP) revealed an enhancement of the nerve signal in both treated groups when compared to the 5% dextrose mice (Figures 5D and 5E). These results proved that the method of nanoprecipitation has no effect on the activity of the siRNA PMP22-SQ NPs.
Figure 5: In vivo efficacity of both siRNA PMP22-SQ NPs formulation. (A). Represents the time taken by the mice of the different treatment groups
to perform the locotronic test. (B and C) Represent the force in gram done by the mice on their forelimb (B) and total limbs (C). (D and E) Show
the compound muscle action potential and nerve signal of the different mice groups respectively. Data represents Mean ± SD. *p<0.05; **p<0.01,
***p<0.001 using one-way ANOVA followed by Bonferroni multiple comparison test. Note:  Wild-type B6,
Wild-type B6,  C61 treated with 5% dextrose,
C61 treated with 5% dextrose,  C61 treated with siRNA PMP22-SQ NPs Forward Nanoprecipitation (FN),
C61 treated with siRNA PMP22-SQ NPs Forward Nanoprecipitation (FN),  C61 treated with siRNA PMP22-SQ NPs Inverse Nanoprecipitation (IN).
C61 treated with siRNA PMP22-SQ NPs Inverse Nanoprecipitation (IN).
Previous successful delivery of squalenoyl siRNA nanoparticles to cancer cells with fusion oncogenes [13], and to Schwann cells in the inherited peripheral neuropathy CMT1A [14], suggest the possibility of applying this therapeutic approach to humans. However, in order to proceed with human clinical trials, the production of squalenoylated siRNA nanoparticles should be optimized by increasing the yield and ensuring the homogeneity of their physicochemical properties.
In all our previous work, the conjugation of siRNA to SQ was achieved at the 10 nmol scale and the nanoprecipitation at 10 μM [13-15]. We observed a sharp decrease in the conjugation yield upon a ten-fold increase in the quantity of conjugation. This was paralleled by an increase in the size and the polydispersity index after nanoprecipitation. To counteract these drawbacks, we reduced the amount of N3-SQ used in the original conjugation step by 50% making it now possible to synthesize 50-times more siRNA-SQ bioconjugate (500 nmol of siRNA-SQ vs. 10 nmol). Since the size and the polydispersity index of siRNA nanoparticles are important for drug delivery a new method of nanoprecipitation named “inverse” was compared to the old method named “forward” that was previously developed in our laboratory [28].
It was found that the inverse method gave smaller nanoparticles with a size of around 100 nm together with a smaller polydispersity index that are suitable for intravenous injection and stable up to one year when stored at 4°C. However, at room temperature they were stable only up to 13 days before the appearance of molds in the solution. This can be overcome under sterile GMP conditions.
The optimal storage conditions of the siRNA-SQ NPs are thus an important criterion for maintaining the stability of the siRNA nanoparticles.
The MD simulations conducted in this work provided further important insight into the molecular details of nanoparticle structure and formation in different solvents. We showed that the early stages of nanoparticle formation, which are accessible in MD simulations, consist of two steps. In the first step, the micellar clusters of conjugates are formed with SQ moieties aggregated together in their center and the RNA helices exposed to the solvent. In the second step, these nano assemblies combine into larger aggregates by means of RNA-RNA interactions. As a result, the hydrophobic mismatch is minimized by hiding highly hydrophobic SQ inside the nano assemblies, while the particle aggregates into a superstructure, which is likely to fill the nanoparticle interior. Both these steps depend on the solvent. In the “inverse” method where water is mainly the solvent at the onset of the nanoprecipitation, the nano assemblies themselves are much smaller and their aggregates are unstable and transient. There is also a significant number of freely diffusing monomers and dimers. This also explains the small size of the siRNA-SQ NPs obtained by DLS measurements and TEM figures. In contrast, in acetone which is the main solvent for the “forward” method of nanoprecipitation, the nano assemblies are much larger and very stable. Such particles accumulate all available conjugate molecules, so there are no free monomers or dimers in the system. The results of the MD simulation confirm the trend in NPs size differences obtained by DLS and TEM. Moreover, although the kinetics of cluster formation is concentrationdependent, the final state of the system depends on the solvent rather than on the initial concentration of conjugate monomers.
Indeed, the clusters are formed faster for a larger concentration of conjugates in the simulation box but their final size is dictated by the solvent polarity.
MD simulations also provide insight into the role of spacer structural rigidity and polarity in the aggregation process. In water, the hydrophobic mismatch between the spacer and water is large enough to favor its attachment to the RNA major groove, which is more hydrophobic than the solvent. The high rigidity of the spacer prevents it from being folded compactly on itself, as it happens with squalene. As a result, there is no gap between squalene and RNA moieties, which imposes steric constraints and only allows the formation of small clusters with only 2-3 conjugates in water. In contrast, the acetone is hydrophobic enough to favor full solvation of the spacer which results in its extended stem-like conformation. This provides a lot of space and flexibility for squalene moieties of different conjugates to interact and allows the formation of a large cluster with up to 8-9 conjugates. This shows that the high rigidity and moderate hydrophobicity of the spacer favor acetone over water for optimal nanoparticle formation.
After studying the physicochemical characteristics, stability, and structure of both nano formulations, it was important to assess their activity in vitro and then in vivo. Based on our previously published data that showed siRNA PMP22-SQ NPs could be a potent therapy for the most common peripheral inherited neuropathy CMT1A [14], we tested the two different siRNA PMP22-SQ NPs formulations on MSC 80 cells and showed a 50% inhibition by both siRNA PMP22-SQ NPs formulation like the naked siRNA. This confirms our previously published data where the modification or the conjugation reaction to SQ didn’t alter the gene inhibition effect of the siRNA [13,14]. siRNA PMP22-SQ NPs prepared by either “forward” or “inverse” methods improved locomotion, muscular strength and the sciatic nerve signal when tested on the C61 CMT1A mouse model in vivo. Altogether these data prove that siRNA-SQ NPs prepared by either of the nanoprecipitation techniques are equally active despite their size difference. We hypothesize that the size of siRNA-SQ NPs does not matter because they interact equally well with serum albumin and LDLs in the blood, which protects them and facilitates their transport to the target tissue [15].
Our results showed that the nanoparticles obtained are strongly dependent on the solvent used. However, the activity of the nanoparticles was not influenced both in vitro and in vivo. Our results allow us to conclude that the protocols established in this work are efficient and reliable for siRNA-SQ nanoparticle production. It opens up the perspective of their therapeutic usage in humans. We believe that the siRNA PMP22-SQ NPs will not trigger any toxicity since the PMP22 gene is specific to the Schwann cells and the squalene is a biodegradable molecule. However, the toxicity of siRNA-SQ NPs should be further investigated.
Suzan Boutary designed and performed the experiments and wrote the manuscript, Guy Khalaf and Marie Caillaud contributed to the synthesis and characterization of the siRNA-SQ NPs, Didier Desmaële developed the azido-squalene synthesis, Semen Yesylevskyy and Christophe Ramseyer performed the molecular modeling of the siRNA-squalene nanoparticles, and, Liliane Massaad-Massade is the PI who supervised and designed the study. All authors contributed to the writing of the manuscript.
The author declared no conflict of interest.
This work was supported by the “SATT Paris Saclay”, program Maturation siCMT and by Fondation NRJ-Institut de France.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Boutary S, Khalaf G, Caillaud M, Desmaële D, Yesylevskyy S, Ramseyer C, et al (2023) Optimizing Structure and Activity of the Squalenoyl- siRNA Nanoparticles. J Clin Toxicol. 13:530.
Received: 20-Feb-2023, Manuscript No. JCT-23-21837; Editor assigned: 22-Feb-2023, Pre QC No. JCT-23-21837 (PQ); Reviewed: 08-Mar-2023, QC No. JCT-23-21837; Revised: 15-Mar-2023, Manuscript No. JCT-23-21837 (R); Published: 22-Mar-2023 , DOI: 10.35248/2161-0495.23.13.530
Copyright: © 2023 Boutary S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.