
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research - (2022)Volume 5, Issue 8
Background: Osteopontin (OPN) is a glycophosphoprotein produced by a variety of cells and has several important physiologic and pathologic roles including cancer pathogenesis through various signaling pathways. Genetic polymorphisms of OPN in 3'UTR and exon may be implicated in the carcinogenesis and progression of colonic carcinomas.
Objectives: This study aimed to find whether OPN rs9138 and rs1126616 single nucleotide polymorphisms were associated with increased risk and progression of Colorectal Carcinoma (CRC).
Subjects and methods: Randomized case control study conducted on 100 CRC patients and 100 apparently healthy subjects. All subjects were investigated for OPN rs9138 and rs1126616 genotyping and CEA, CA 19-9 and OPN plasma levels. The genotypes were assayed using Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) while tumor markers serum levels were measured by ELISA.
Results: The results revealed that AC genotype of rs9138 and CC and CT genotype of rs1126616 were associated with increased risk of CRC. The C allele of both rs9138, rs1126616, and the haplotypes C (rs1126616)- C (rs9138) and C (rs1126616)- A (rs9138) were associated with increased CRC risk. Serum OPN protein expression in CRC patients was significantly increased as compared to healthy controls and related to severity of the cancer.
Conclusion: The OPN rs9138 and rs1126616 gene polymorphism were associated with increased CRC risk and the OPN serum level could be used as a possible diagnostic and prognostic marker of CRC.
Osteopontin (OPN); Gene polymorphism; Colorectal carcinoma; Malignant tumors
Malignant tumors of the colon and rectum are considered the most well-known tumors in the world and are the 4th leading cause of cancer death after lung, breast and cervical cancers [1]. According to the etiology, 75% of colonic cancers are of sporadic origin, and 25% are due to hereditary lesion [2].
CRC develops through a sequential accumulation of genetic mutations. Recurrence and mortality rates of CRC depend on stage of the disease at which diagnosis is made. Early diagnosis of CRC through mass screening programs can diminish the risk of CRC mortality [3].
Non-invasive tests used for screening of CRC are Fecal Occult Blood Test (FOBT) [4], Carcino-Embryonic Antigen (CEA), [5] and Carbohydrate Antigen (CA) 19-9 [3]. The definitive method for diagnosis of colonic cancers is endoscopy [6], but due to its high costs and inconvenience, its utilization is limited [7].
OPN is a glycophosphoprotein produced by a variety of cells [8] and has several important physiologic and pathologic roles, as bone turnover, wound healing, inflammation, autoimmune diseases and cancer pathogenesis by enhancement of various signaling pathways via attachment to surface receptors as integrin's and CD44 variants [9].
There are few studies about the role of OPN gene in development and progression of CRC [10].Genetic polymorphisms of OPN in 3'UTR and exon may be implicated in the carcinogenesis and progression of colonic carcinomas [11].
The aim of this study is to determine whether OPN rs9138, rs1126616 polymorphisms is associated with increased the risk and progression of colorectal tumors. Also, assessment of serum OPN, CEA, and CA 19-9 levels in CRC patients and their relation with different genotypes.
Subjects and methods
Randomized case control study was performed on 200 subjects who were divided into two groups. Group 1 included 100 biopsy confirmed CRC patients with no other tumors; they were selected from the Gastroenterology surgical center, Mansoura University. According to the AJCC staging 8th edition (2017), staging of the CRC was executed and patients were divided into early-stage group (IandII) that included 61 patients and late-stage group (IIIandIV) that included 39 patients. The clinical and pathological data were collected from patients' records. Group 2 included 100 apparently healthy age and sex matched subjects.
Sample collection: Venous blood sample was taken and divided into 2 tubes: a plain sterile vacutainer for serum separation and assayed for CEA, CA 19.9 land Osteopontin evels and a sterile EDTA vacutainer for the OPN genotyping.
Genetic analysis of OPN polymorphism: Thermo Scientific Gene JET whole Blood Genomic DNA Purification Mini Kits was used for the extraction of genomic DNA from peripheral blood leukocytes. Genotyping of OPN rs1126616 and rs9138 were performed by PCR-restriction fragment length polymorphism technique. PCR amplification was performed according to Fan et al. [10] study (Table 1).
| SNP | Primers (Bio Basic Canada Inc.) | Annealing temperature | Restriction enzyme | |
|---|---|---|---|---|
| For OPN rs9138 | Forward | 5'TGGTTGTAGACCCCAAAAGTA3 | 56°C | AccI |
| Reverse | 5'AACCGTGGGAAAACAAATAA3' | |||
| For OPN rs1126616 | Forward | 5'CCGTGGGAAGGACAGTTATG 3' | 55°C | AluI |
| Reverse | 5'TTTAATTGACCTCAGAAGATGCAC' | |||
Table 1: Primers, restriction enzymes, annealing temperature of OPN SNPS.
The amplified PCR products were digested by appropriate restriction enzyme. AccI restriction enzyme (Thermo-scientific fast Digest enzyme) is used for OPN rs9138 and the PCR products were divided into two fragments (382 bp and 186 bp) after incubation for about 14 h at 37°C. Regarding SNP of OPN rs1126616, the PCR products were divided into two fragments (198 bp and 92 bp) after being digested by AluI restriction enzyme (Thermo-scientific fast Digest enzyme) for about 13 h at 37oC. Separation of PCR products digested by restriction enzyme was performed by using 2% agarose gel electrophoresis (Figures 1 and 2).
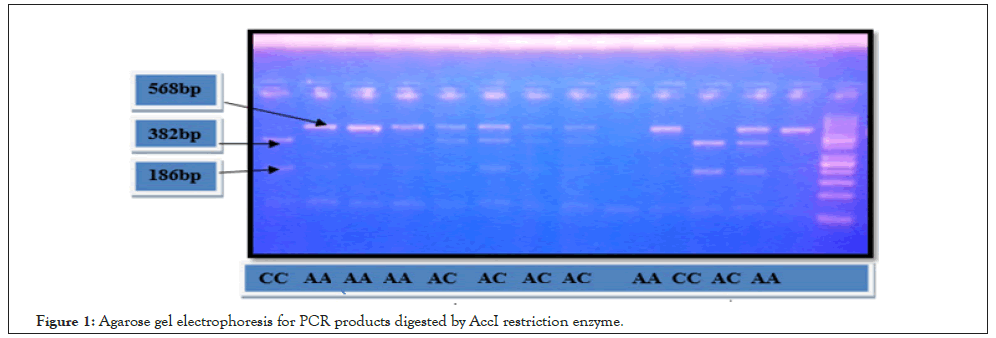
Figure 1: Agarose gel electrophoresis for PCR products digested by AccI restriction enzyme.
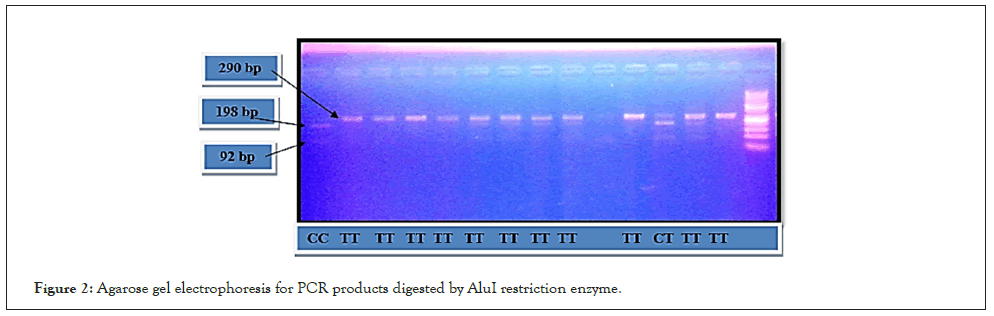
Figure 2: Agarose gel electrophoresis for PCR products digested by AluI restriction enzyme.
Serum OPN protein levels and tumor markers assay: The serum OPN levels were determined by Enzyme-Linked Immunosorbent Assay (ELISA) by protocol offered from the manufacturer SunRed, China. The optical density was detected at 450nm using STAT Fax ELISA plate reader.
Statistical analysis
The statistical analysis of these data was executed by usage of excel program (Microsoft Office 2007) and Statistical Package for Social Science program (IBM SPSS) (SPSS, Inc, Chicago, IL) version 20. Qualitative data were illustrated as frequencies and percentages and Chi square test was chosen to compare different groups. Quantitative variables were summarized as mean ±SD for parametric values, and median, range for nonparametric ones.
Comparisons between two studied groups were performed by utilization of t-test (parametric data) or Man Whitney U test (nonparametric data). One way ANOVA test accomplished to compare between multiple groups. Diagnostic efficacy of the studied tumor markers was analyzed by plotting a ROC curve and AUC of studied markers either used alone or in combination was calculated. By these curves, the optimal best cut-off values with highest sensitivities, specificities were obtained. Pearson's correlation coefficient was performed to detect the correlation between parameters. Odds ratio and 95% CI were calculated. The logistic regression analysis was performed for prediction of CRC risk. The haplotype frequencies were examined by Haploview program. p is significant if <0.05 at confidence interval 95%.
Demographic data of the subjects and the clinic-pathological characteristics and laboratory da-ta were summarized in Table 2. There was no statistically significant difference between cases and controls as regards gender and age. This indicated that these variables were chosen adequately and appropriately. Serum CEA, CA 19-9 and OPN levels were significantly increased CRC patients than control (p=0.0001). Serum OPN level only showed significant increase in late stages of CRC than early ones (p=0.0001) (Figure 3), while there was no significant difference between early and late stages of CRC patients regarding CEA and CA 19-9 (P=0.22; 0.8), respectively. There was no significant correlation between plasma OPN protein and CEA or CA 19–9 (R=0.002, 0.97, p=-0.07, 0.45), respectively (Table 2).
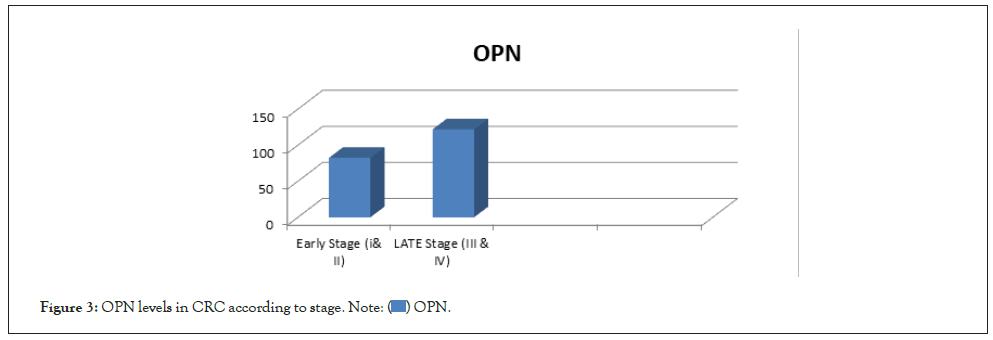
Figure 3: OPN levels in CRC according to stage.
| Age (years) | Mean ±SD | Case | Control | P value |
|---|---|---|---|---|
| 50.4 ± 11 | 47.7 ± 10 | 0.07 | ||
| Gender No. (%) | Male | 59 (59%) | 51(51%) | 0.25 |
| Female | 41 (41%) | 49 (49%) | ||
| Tumor site No. (%) | Colon | 51 (51%) | ||
| Rectum | 49 (49%) | |||
| Tumor stage No. (%) | Early (I and II) | 61 (61%) | ||
| Late (III and IV) | 39 (39%) | |||
| Pathologic type No. (%) | Adenocarcinoma | 90 (90%) | ||
| Mucinous adenocarcinoma | 10 (10%) | |||
| CEA ng/ml | Median(range) | 6.05 (0.6-308) | 1.9 (1-3.5) | 0.0001 |
| CA 19-9 IU/ml | Median(range) | 33.5 (8-101) | 19 (2-31) | 0.001 |
| OPN ng/ml | Median(range) | 91 (16-613) | 39 (8-101) | 0.001 |
Table 2: Clinic-pathologic and laboratory data of studied groups.
Through ROC curve, OPN yielded the best AUC than CEA and CA19-9. The optimal cut off value of OPN with highest sensitivity and specificity for screening of CRC was 59.5 ng/ml. Combined analysis of CEA+CA19-9+OPN yielded highest AUC than the use of best single tumor marker; OPN, with diagnostic efficacy 95.1% (Figures 4 and 5).
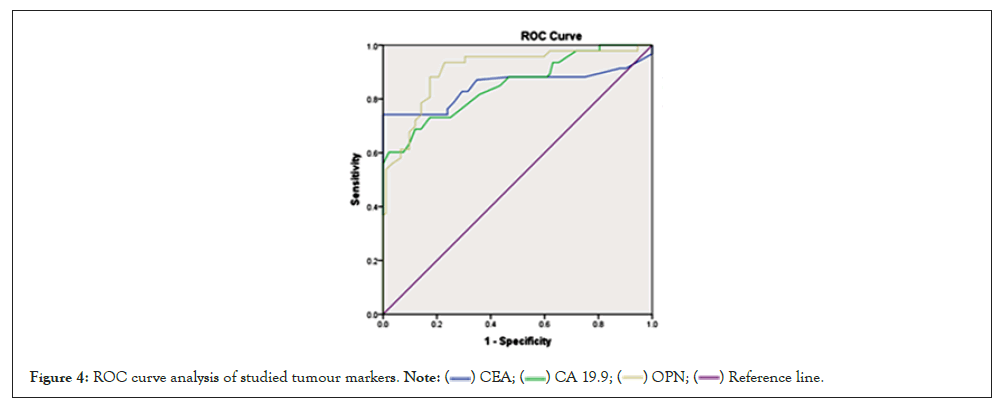
Figure 4: ROC curve analysis of studied tumour markers.
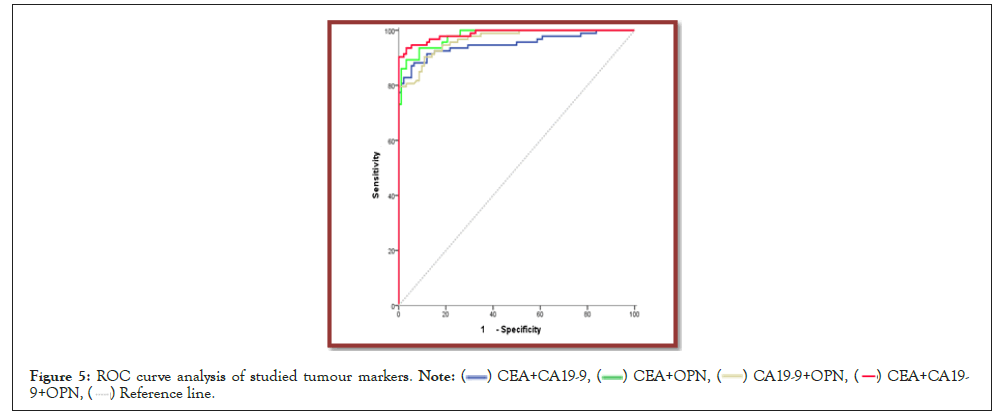
Figure 5: ROC curve analysis of studied tumour markers.
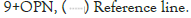
Regarding OPN rs9138, AC and CC genotypes were significantly higher in CRC patients versus control subjects (p2<0.01 and P3<0.01respectively), while AA genotype of OPN rs9138 is significantly lower in CRC patients when compared to control group. The analysis of frequency of OPN rs9138 alleles (A and C) showed statistically significant difference between CRC patients and control group; C allele was significantly higher in CRC patients (42.5%) versus control subjects (26%), (p4<0.001, OR=2.10, 95% CI=1.35-3.28). While T allele was significantly lower in CRC patients (57.5%) versus controls (74%), (p5=0.001, OR=0.48, 95% CI=0.30-0.74) (Table 3).
| SNP | Genotype/Allele | CRC N=100 (%) | Control N=100 (%) | OR (CI 95%) | P value |
|---|---|---|---|---|---|
| rs9138 | AA | 31 (31%) | 55 (55%) | 0.37(0.2-0.6) | <0.001 |
| AC | 53 (53%) | 38(38%) | 1.84(1.01-3.63) | 0.01 | |
| CC | 16 (16%) | 7 (7%) | 2.53(2.53-7.18) | 0.01 | |
| A | 115 (57.5%) | 148 (74%) | 2.10(1.35-3.28) | 0.001 | |
| C | 85 (42.5%) | 52 (26%) | 0.48(0.30-0.74) | <0.001 | |
| rs1126616 | TT | 21 (21%) | 85 (85%) | 0.05(0.02-0.1) | 0.001 |
| CT | 75 (75%) | 15 (15%) | 17(2.37-5.27) | 0.001 | |
| CC | 4 (4%) | 0 (0%) | Not calculated | 0.059 | |
| T | 117 (58.5%) | 185(92.5%) | 0.10(0.05-0.19) | 0.001 | |
| C | 83 (41.5%) | 15 (7.5%) | 9.69(5.18-18.19) | 0.001 |
Table 3: Genotype and allele distributions of OPN polymorphisms in cases and controls.
CT genotype of OPN rs1126616 was significantly higher in CRC patients when compared to control subjects (p2=0.001, odds ratio 17, 95%CI 2.37-5.27), while CC genotype showed non-significant elevation in CRC patients when compared to control subjects (p3= 0.059, odds ratio and 95%CI can't be calculated). On the other hand, TT genotype was significantly higher in control healthy subjects (p1=0.001, odds ratio 0.05, 95%CI 0.02- when compared to CRC group (Table 3).
The analysis of frequency of OPN rs1126616 alleles (T and C) showed statistically significant difference between CRC patients and control group; C allele was significantly higher in CRC patients (41.5%) versus control subjects (7.5%), (p4= 0.001, OR=9.69, 95% CI=5.18-18.19). While T allele was significantly lower in CRC patients (58.5%) versus controls (92.5%), (p5=0.001, OR=0.10, 95% CI=0.05-0.19) (Table 3).
Haplotype analysis revealed that TA showed the highest frequencies in cases and controls, while CC showed the lowest frequency in controls, and CA had the lowest frequencies in cases. TA and TC haplotypes showed protective effects against CRC development. While, CC and CA haplotypes were considered risky haplotypes for CRC development within healthy control subjects (Table 4).
| Haplotype rs1126616- rs9138 | Total frequency | Control frequency | CRC frequency | OR (95% CI) | P value | ||
|---|---|---|---|---|---|---|---|
| TA | 0.561 | 0.685 | 0.437 | 0.357 | 0.297 | 0.429 | <0.001 |
| TC | 0.194 | 0.24 | 0.148 | 0.55 | 0.438 | 0.69 | 0.039 |
| CC | 0.149 | 0.02 | 0.277 | 1.873 | 1.605 | 9.853 | <0.001 |
| CA | 0.096 | 0.055 | 0.138 | 2.751 | 1.986 | 3.81 | 0.006 |
Table 4: Comparison of OPN haplotype frequencies and risk of CRC within healthy control subjects.
Regression analysis was conducted for prediction of CRC within healthy control subjects, using age; gender CEA, CA19-9, OPN, rs1126616, rs9138 as covariates in Table 4. Higher CEA, CA19- 9, OPN, rs1126616 and rs9138 were associated with higher risk of CRC in univariate analysis. Those covariates which were significant in univariate analysis were introduced into multivariate analysis which reveal that higher CEA, CA19-9, OPN, rs1126616 were considered as predictors of CRC development within healthy control group (Table 5).
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| p | OR | 95% CI | p | OR | 95% CI | |||
| Age | 0.071 | 1.049 | 0.02 | 1.08 | ||||
| Male | 0.256 | 1.383 | 0.791 | 2.418 | ||||
| CEA | <0.001 | 2.422 | 1.796 | 3.267 | 0.048 | 1.855 | 1.006 | 3.423 |
| CA19-9 | <0.001 | 1.145 | 1.101 | 1.19 | 0.002 | 1.197 | 1.066 | 1.344 |
| OPN | <0.001 | 1.071 | 1.05 | 1.092 | 0.001 | 1.092 | 1.036 | 1.15 |
| rs1126616 (CT+CC) | <0.001 | 2.317 | 1.273 | 14.23 | 0.001 | 2.914 | 1.915 | 16.692 |
| rs9138 (AC+CC) | 0.001 | 2.72 | 1.525 | 4.852 | 0.272 | 2.863 | 0.438 | 18.733 |
Table 5: Regression analysis for prediction of CRC within healthy control subjects.
OPN was one of the principal genes involved in the development and carcinogenesis of CRC [12]. The OPN protein is regulated by transcription factors and genetic polymorphisms affecting 3'UTR [13], exons [14], and the promoter region [15]. Fan et al., [10] revealed that the OPN gene polymorphisms increased the risk to CRC development.
Regarding rs1126616 (+750C/T, exon 7) SNP, it was observed that CT genotype was significantly associated with increased risk of CRC. No significant elevation of CC genotype was observed in CRC patients versus control group. Individuals carrying C allele of rs1126616 was more vulnerable to CRC than T allele carriers (Table 3). These findings are supported partially by Fan et al., [10] who revealed CC and CT genotypes of rs1126616 were associated with increased CRC risk.
Melanitou [16] observed that the frequency of the combined genotypes (CT+CC) frequencies was significantly higher in CRC patients than in controls as compared to the frequency of TT genotypes and agreed with this study's results in that C allele was significantly higher in CRC patients than controls.
The genotypes CC and AC of rs9138 (+1239A/C, 3’UTR) were significantly associated with increased risk of CRC and the patients carrying the C allele (rs9138) have a significantly higher risk for developing CRC (Table 3). This suggests that the carriers of this allele may be more prone to CRC development. These results came in partial accordance with Fan et al., [10], who observed in that the genotypes AA and AC of rs9138 were associated with increased risk of CRC as compared with the CC genotype. On the other hand, [16] found no significant difference by comparing the genotype and allele frequencies of OPN rs9138 in the two studied groups, but observed in female, the frequencies of the combined genotypes (AC+CC) and C allele were significantly higher in CRC patients than those of controls. This controversy between results may be due to differences in sample size and larger studies are needed to confirm these association.
To predict CRC within healthy control subjects, regression analysis were performed in Table 5. It was observed in univariate analysis that higher CEA, CA19-9, OPN, rs1126616 and rs9138 were associated with increased risk of CRC. While in covariates analysis, higher CEA, CA19-9, OPN, rs1126616 only were considered as predictors of CRC development within healthy control group.
The polymorphism of rs1126616 was perfect LD with the polymorphism of rs9138, so we observed that TA and TC haplotypes have protective effects against CRC development. While, CC and CA haplotypes were considered as risky haplotypes for CRC development within healthy control subjects (Table 4). These findings agreed with Melanitou, et al., [16], who reported that patients carrying the haplotype of C (rs1126616) –C (rs9138) had a significant higher risk for CRC development. Similarly, Fan et al., [10] reported that haplotype of C (rs1126616)- A (rs9138) had higher risk for CRC.
In the present study, there were no correlation between OPN rs9138 and OPN rs1126616 genotypes in Table 5 with clinical, pathological and laboratory data of patients. This was consistent with Fan et al., [10] who found that the site, histological type, differentiation degree, and stage of the tumor were not associated with OPN SNPs and also, plasma lipid levels, tumor markers (CEA and CA19-9) showed no statistical significant difference with OPN SNPs.
However, Melanitou et al., [16] found that OPN rs1126616 was more common in the old-age CRC group (>40years) but not in the young-age group (<40years) as compared to healthy subjects. As regards OPN rs9138 polymorphism, Melanitou Kamal et al., [16] reported that the mutant C allele is predominant in female CRC patients (50.9%) than females of healthy group (36.2%), suggesting that females having the mutant OPN rs9138 C allele had increased risk of CRC.
Despite, the correlations between the rs9138 and rs1126616 SNPs and the increased risk of CRC, the OPN levels in our study didn't show any significant differences between various genotypes. This came in accordance with Fan et al., [10] and Melanitou et al., [16] who observed no correlation be-tween OPN genotypes and OPN serum concentrations.
Through ROC curve, OPN yielded the best AUC than CEA and CA 19-9. On combined analysis of studied tumor markers OPN+CEA+CA 19-9, the AUC, sensitivity and specificity had been in-creased dramatically and diagnostic efficacy approached 95.1%. So, this study concluded that com-bined OPN+CEA+CA 19-9 analysis can significantly increase the detection rate of CRC and decrease the need for the invasive methods for CRC screening as colonoscopy. These results were in agreement with Fan et al., [10], Catalán et al., [17] and Melanitou et al., [16] studies which reported higher AUC of plasma OPN than CEA and CA19-9. Therefore, this indicates that serum OPN has diagnostic efficacy for CRC than CEA and CA19-9.
The results of this study might improve the understanding about the genetic changes in the CRC; and identification of diagnostic, prognostic, and predictive markers that support CRC prevention, early detection, and treatment. This study concluded that the AC and CC genotypes of rs9138 and CT genotype of rs1126612 genotypes were associated with the risk of developing colorectal cancer in Egyptian subjects. Also, carriers of C allele of rs9138 and C allele of rs112612 are at increased risk of CRC. Also, our study concluded that simultaneously testing OPN+CEA+CA19-9 can improve safety and increase diagnostic sensitivity in identifying people with CRC.
The authors have no competing interests to declare that are relevant to the content of this article.
This study was approved by ERP department, faculty of medicine, Mansoura University and written informed consent was obtained from each participant.
We would like to thank all members in Clinical Pathology department, faculty of Medicine, Mansoura university and all our colleges who helping us to complete this work.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
[Crossref] [Google Scholar] [PubMed].
Citation: Rizk E, El-Arman M, El-Baiomy A, Kandil T, Mourad S, Elmam O (2022) Osteopontin Rs9138 and Rs1126616 Gene Polymorphism in Egyptian Patients with Colorectal Carcinoma. J Clin Chem Lab Med. 5:238.
Received: 30-Aug-2022, Manuscript No. JCCLM-22-19046; Editor assigned: 02-Sep-2022, Pre QC No. JCCLM-22-19046 (PQ); Reviewed: 16-Sep-2022, QC No. JCCLM-22-19046; Revised: 23-Sep-2022, Manuscript No. JCCLM-22-19046 (R); Published: 30-Sep-2022 , DOI: 10.35248/JCCLM.22.5.238
Copyright: © 2022 Rizk E, et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.