Clinical Pediatrics: Open Access
Open Access
ISSN: 2572-0775
ISSN: 2572-0775
Research Article - (2024)Volume 9, Issue 4
Background: The prevalence and antibiogram of pathogenic E. coli strains which cause diarrhea vary from region to region, and even within countries in the same geographical area. In Ethiopia, diagnostic approaches to E. coli induced diarrhea in children less than five years of age are not standardized. The aim of this study was to determine the involvement of pathogenic E. coli strains in child diarrhea and determine the antibiograms of the isolates in children less than 5 years of age with diarrhea at Addis Ababa university college of health sciences Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia.
Methods: A purposive study which included 98 diarrheic children less than five years of age was conducted at Addis Ababa university college of health sciences, Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia to detect pathogenic E. coli biotypes. Stool culture was used to identify presumptive E. coli isolates. Presumptive isolates were confirmed by biochemical tests and anti-microbial susceptibility tests were performed on confirmed E. coli isolates by disk diffusion method. DNA was extracted from confirmed isolates by heating method and subjected to PCR for the presence of virulence genes. Amplified PCR products were analyzed by agarose gel electrophoresis. Data were collected on child demographics and clinical conditions using administered questionnaires. The prevalence of E. coli strains from the total diarrheic children and the prevalence of pathogenic strains from total E. coli isolates along with their susceptibility profiles; the distribution of pathogenic E. coli biotypes among different age groups and between the sexes were determined by using descriptive statistics.
Results: Out of 98 stool specimens collected from diarrheic children less than 5 years of age, 75 presumptive E. coli isolates were identified by culture; further confirmation by biochemical tests showed that only 56 of the isolates were E. coli; 29 of the isolates were found in male children and 27 of them in female children. Out of the 58 isolates of E. coli, 25 pathotypes belonging to different classes of pathogenic strains: STEC, EPEC, EHEC, EAEC were detected by using PCR technique. Pathogenic E. coli exhibited high rates of antibiotic resistance to many of the antibiotics tested. Moreover, they exhibited multiple drug resistance.
Conclusion: This study found that the isolation rate of E. coli and the involvement of antibiotic resistant pathogenic E. coli in diarrheic children are prominent and hence focus should be given on the diagnosis and antimicrobial sensitivity testing of pathogenic E. coli at Addis Ababa university college of health sciences Tikur Anbessa specialized hospital. Among antibiotics tested, cefotitan could be a drug of choice to treat E. coli.
E. coli; Pathogenic; Diarrhea; Children; Antibiotic susceptibility
EAEC: Entero Aggregative E. coli; EHEC: Entero Hemorrhagic Escherichia coli, STEC: Shigatoxin producing Escherichia coli; EPEC: Entero Pathogenic Escherichia coli; HGT: Horizontal Gene Transfer; EMB: Eosin Methylene Blue; BFP: Bundle Forming Pilus; MMR: Methyl Directed Mismatch Repair; SD: Standard Deviation; WHO: World Health Organization; PCR: Polymerase Chain Reaction; MDR: Multi Drug Resistance; DNA: Deoxyribonucleic Acid
E. coli is a gram negative, rod shaped, facultative anaerobe bacterium that belongs to the family Enterobacteriaceae. It is normally found in the intestines of humans and is involved in the maintenance of the balance of the intestinal microbiota. The E. coli genome has a single circular chromosome with double stranded DNA. Some strains may also contain extra chromosomal genetic elements such as plasmids. E. coli can be easily genetically manipulated and readily grows in the laboratory. Therefore, E. coli has always been a common and suitable model organism for different genetic studies.
The reference E. coli genome (k-12) has 4288 protein coding genes. Genome plasticity through horizontal gene transfer is high in E. coli as indicated by the presence of insertion sequences, phage remnants and additional sequences of unusual composition in the genome [1].
Although most E. coli are commensals, some strains can be pathogenic due to additional virulence genes found in chromosomes and/or plasmids. Virulence genes can be generated by mutation; they can also be acquired from mobile genetic elements through horizontal gene transfer. Clusters of virulence genes can be found on plasmids or integrated into the chromosome in pathogenic strains. These clusters of genes also called pathogenicity islands are usually flanked by mobile genetic elements such as bacteriophages, insertion sequences, etc. and often insert near transfer RNA genes [2].
E. coli bacteria evolve from distinctive clonal groups and most strains of E. coli (85%-90%) can be assigned by simple PCR technique into different phylogenetic groups based on the presence/absence of the genes designated as chuA and yjaA and a DNA fragment designated as TSPE4.C2. Other more complex methods of phylogenetic grouping are enzyme electrophoresis and ribotyping. There are four known phylogenetic groups (A, B1, B2, D) of E. coli among which groups A and B1 have been found to be associated with intestinal human and mammalian pathogenicity [3].
A study on mutation rates of E. coli has shown that mutation rates in isolated pathogenic E. coli are higher than the average bacterial population with defect in methyl directed mismatch repair being the predominant underlying cause. However, high mutation rates due tomethyl directed mismatch repair were demonstrated in both pathogenic and commensal E. coli strains in another independent survey of mutation rates of E. coli populations isolated from distinct environments. Genome size of E. coli markedly varies between commensal and pathogenic strains and the extra genetic material in pathogenic strains can contain either virulence or fitness genes.
The overall target genes for detection of pathogenic E. coli are varied between different studies. Pathogenic E. coli strains can be detected by Polymerase Chain Reaction (PCR) which is a powerful molecular biology technique that can detect target DNA of many kinds of pathogens in various clinical specimens. PCR is preferred over other methods of detection because it gives rapid, reliable results with greater specificity and sensitivity [4].
Based on unique sets of virulence and colonization factors encoded in the chromosomal or episomal structures and target genes, pathogenic E. coli bacteria can be grouped in to different strains. Enteroinvasive E. coli strains (EIEC) possess invasion genes virF and ipaH; Entero Pathogenic E. coli strains (EPEC) possess a pathogenicity island in their genome called Locus of Enterocyte Effacement (LEE) which encodes for the target gene intimin, designated as EAE [5].
These strains are also called attaching/effacing E. coli because they attach intimately and efface cytoplasm and microvilli from the intestinal epithelial cells of their hosts. Typical Enteropathogenic E. coli contains EPEC Adherence Factor (EAF) plasmid which codes for the target gene Bundle Forming Pillus (bfp) in addition to LEE. Enterotoxigenic E. coli are detected by their heat labile and heat stable enterotoxin genes (LT, ST); shiga toxin producing E. coli also called verotoxin producing E. coli possess shiga toxin genes (stx1 and stx2); entero aggregative E. coli produce pic and other toxins.
Diarrhea is a clinical condition characterized by frequent bowel movement with loose stools and accompanying signs and symptoms like fever and vomiting. Acute diarrhea is a term used to describe the presence of three or more loose watery stools within 24 hours; dysentery indicates presence of blood and mucus in diarrheal stools; and persistent diarrhea is diarrhea lasting for more than 14 days. Children in developing countries get exposed to many bacterial enteric pathogens at very early age and suffer many episodes of diarrhea as a result; pathogenic E. coli is among these enteric pathogens. Pathogenic E. coli has been associated with diarrheal disease in different parts of Africa particularly among young children, HIV positive and visitors from abroad [6].
The rational management of infectious diarrhea requires highly selective use of laboratory tests for these varied etiologic agents, depending on the clinical and epidemiologic setting; information generated from the study of pathogenic E. coli permits a practical approach to the diagnosis and management of diarrhea [7].
Study area
This study was conducted at Tikur Anbessa specialized hospital, Addis Ababa university college of health sciences, Addis Ababa. Addis Ababa is the capital and largest city of Ethiopia, located on a well-watered plateau surrounded by hills and mountains at an altitude of about 2500 m above sea level.
The average annual temperature and rainfall are 21℃ and 1800 mm, respectively. Tikur Anbessa hospital is the largest hospital in the country and functions under the authority of the Addis Ababa university college of health sciences.
Study population
Patients included in this study were children under 5 years old with 3 or more loose stools in 24 hours or with an episode of bloody diarrhea. Children that received previous antimicrobial drug treatment were excluded from this study [8].
Study design and sampling methodology
The study was purposive type, i.e., samples were collected from all children less than five years of age with diarrhea. Sample collection was done from January 2017 to March 2018. Approximately 50 gm of feces was collected from children with diarrhea in accordance with standard laboratory specimen collection procedures. Specimens from diarrheic children were collected in sample cups on to which buffered peptone water was added for enrichment. Specimens were labeled with unique sample identification numbers, transported in ice box to biomedical laboratory of microbial, cellular and molecular biology at college of natural sciences, Addis Ababa university and inoculated in to primary culture media within the same day of collection.
Isolation of E. coli
Broth specimens were be inoculated on MacConkey agar and incubated aerobically at overnight. Lactose fermenting colonies on MacConkey agar were then sub cultured in to eosin methylene blue and incubated aerobically at overnight. Green metallic sheen colonies on eosin methylene blue were considered as presumptive E. coli isolates.
Presumptive isolates were stored in nutrient broth for further identification by biochemical tests. All the isolates were also stained by gram stain to determine cell morphology and purity of the isolates [9].
Biochemical characterization of E. coli isolates
Presumptive E. coli isolates were further characterized for their biochemical activity using the biochemical tests Indole, Methyl red, Vogues Proskuer and Citrate utilization (IMViC). Bacterial isolates that exhibited IMViC pattern of (+ + - -) respectively were considered as E. coli isolates.
Indole test
A sterilized test tube containing 4 ml of tryptophan broth was inoculated aseptically by taking an inoculum from 18 hrs to 24 hrs culture on EMB. The broth was incubated at 37°C for 24-28 hours. 0.5 ml of Kovac’s reagent was added to the broth culture and the presence or absence of ring was observed. Formation of a pink color in the reagent layer on top of the medium within seconds of adding the reagent was considered as positive result. Absence of ring formation considered as a negative result [10].
Vogues-Proskuer (VP) test
The medium was inoculated with an inoculum taken from an 18 hour-24 hour pure culture and incubated aerobically at 37℃for 24 hours. 1 ml of the broth was transferred to a clean test tube following 24 hours of incubation. The remaining broth was reincubated for an additional 24 hours. 0.6 ml of 5% alphanaphthol was added to the 1 ml broth and next 0.2 ml of 40% KOH was added. By gently shaking to expose the medium to atmospheric oxygen, the tube was allowed to remain undisturbed for 10 minutes-15 minutes. Observation of a pink red color development was considered as a positive VP test. A negative VP test was demonstrated by the appearance of a yellow color on the surface of the medium. Development of a copper like color was also interpreted as negative.
Methyl red test
Following 48 hours of incubation, 2.5 ml of the broth was transferred to a clean test tube. Five drops of methyl red indicator were added. Development of a stable red color on the surface of the medium after the addition of methyl red indicator was interpreted as positive test. A negative methyl red test was demonstrated by the development of a yellow color on the surface of the medium [11].
Citrate utilization test
Simmons citrate agar was inoculated on the slant by touching the tip of a needle to a colony that is 18 hours to 24 hours old and incubated at 37℃ for 24 hours. Development of blue color on the slant surface due to the alkaline carbonates and bicarbonates produced as byproducts of citrate catabolism increasing the pH was considered as positive result. The absence of color change (the medium remains deep green) was considered as negative result.
Antimicrobial sensitivity testing
The antimicrobial susceptibility/resistance profiles of the bacterial isolates were determined using Kirby Bauer disk diffusion method. Disks impregnated with the following antibiotics were used: Trimethoprim (5 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), ampicillin (10 μg), neomycin (10 μg), gentamycin (10 μg), tetracycline (30 μg), compound sulfonamides (300 μg), chloramphenicol (30 μg), cefotetan (30 μg), norfloxacin (10 μg) and streptomycin (25 μg). Pure bacterial colonies were inoculated into 7 ml of tryptophan soya broth and incubated at 37℃ for 18 hours until turbidity is seen and were compared to the 0.5 McFarland standards. Mueller-Hinton agar was used as plating medium. Fifteen minutes after inoculation of the plates using sterile swabs, the antibiotic impregnated disks were applied on the surface of inoculated plates with sterile forceps. All the disks were gently pressed down onto the agar with forceps. The plates were inverted and then incubated aerobically for 18 hours at 37℃ [12]. The diameters of the zones of inhibition were measured to the nearest whole millimeter using the transparent ruler and were interpreted as susceptible, intermediate and resistant based on the recommendations of clinical laboratory standards institute.
Virulence gene detection
DNA extraction: Biochemically confirmed E. coli isolates were grown in nutrient broth at 37℃ overnight. Exactly 1.5 ml of the culture was spun by centrifugation at 5000 g for 10 min.
The bacterial pellet was lysed by adding 50 μl of double distilled water and boiling in a water bath at 95℃ for 10 minutes. The lysate was then centrifuged again as before and 50 μl of the supernatant used directly as template for PCR.
Detection of virulence gene sequences by PCR
After extraction, the bacterial DNA was subjected to PCR for the presence of virulence genes. According to the annealing temperatures of the different primers used, five PCR assays were performed. The PCR experiments were carried out using the following protocols.
To detect the presence of stx2 (shigatoxin) genes of STEC and EHEC, a reaction was set up in a 25 μl reaction volume in a PCR master mix (Himedia; India, 2017) containing 1 μ1 of each primer (EVS1, EVC2), 2.5 μl of PCR buffer with 17.5 mmol of MgCl2, 1 μl of 0.35 mm of each dNTP, 0.5 μ1 of Taq polymerase enzyme, 14 μ1 of double distilled water and 3 μl of template DNA. The reaction mixture was amplified with an initial denaturation of 1 cycle for 3 min. at 95℃; 30 cycles each consisting, 40 s at 95℃, 40 s at 55℃, 30 s at 72℃; and a final extension of 1 cycle for 8 min. at 72℃ [13].
To detect the EAE (intimin) gene of EPEC and EHEC strains, a reaction was set up in a 25 μl reaction tube in a PCR master mix (Himedia; India, 2017) containing 1 μ1 of each primer (EAE1 and EAE2), 2.5 μl of PCR buffer with 17.5 mmol of MgCl2, 1 μl of 0.35 mm of each dNTP, 1 μ1 of 50 Mmol MgCl2, 0.5 μl of Taq polymerase enzyme, 15 μ1 of sterile distilled water and 3 μl of template DNA. The reaction mixture was amplified with an initial denaturation of 1 cycle for 3 min. at 95℃; 35 cycles, each consisting of 40 s at 95℃, 60 s at 55℃ and 60 s at 72℃; and a final extension of 1 cycle for 10 min. at 72℃.
To detect the bfp (bundle forming pillus) gene of typical EPEC strains, a reaction was set up in a 25 μl reaction tube in a PCR master mix (Himedia; India, 2017) containing 0.5 μ1 of each primer (BFPF, BFPR) containing 2.5 μl of PCR buffer with 17.5 mmol of MgCl2, 1 μl of 0.35 mm of each dNTP, 0.3 μl of Taq polymerase enzyme, 16.2 μl of sterile distilled water and 3 μl of template DNA. The reaction mixture was amplified with an initial denaturation of 1 cycle for 3 min. at 95℃; 30 cycles, each consisting of 40 s at 95℃, 40 s at 57℃, 30 s at 72℃; and a final extension of 1 cycle for 8 min at 72℃ [14].
To detect hylA (hemolysin) gene of EHEC, reaction components were mixed in a 25 μl reaction tube in a PCR master mix (Himedia; India, 2012) containing 1 μ1 of each primer (EHECF, EHECR) containing 2.5 μl of PCR buffer with 17.5 mmol of MgCl2, 1 μl of 0.35 mm of each dNTP, 0.3 μl of Taq polymerase enzyme, 16.2 μl of sterile distilled water and 3 μl of template DNA. Then, the reaction mixture was amplified with an initial denaturation of 1 cycle for 3 min. at 95℃; 30 cycles, each consisting of 40 s at 95℃, 1 min. at 45℃, 1 min at 72℃; and a final extension of 1 cycle for 10 min at 72℃ [15].
To detect aatA (antiagrgation transporter gene) of EAEC strain reactions were set up in a 25 μl reaction tube in a PCR master mix containing 1 μ1 of each primer (EAECF, EAECR) containing 2.5 μl of PCR buffer with 17.5 mmol of MgCl2, 1 μl of 0.35 mm of each dNTP, 0.3 μl of Taq polymerase enzyme, 16.2 μl of sterile distilled water and 3 μl of template DNA. The reaction mixture was amplified with an initial denaturation of 1 cycle for 3 min. at 95℃; 30 cycles each consisting of, 40 s at 95℃, 1 min. at 45℃, 30 s at 72℃ ; and a final extension of 1 cycle for 10 min. at 72℃ [16].
All amplifications were carried out in a thermal cycler (applied BiosystemsStepOne™ real-time PCR system thermal cycling block).
Agarose gel electrophoresis
Amplified PCR products were analyzed by agarose gel electrophoresis at 120 volt for 30 minutes in 1.5% agarose containing ethidium bromide (0.5 μg ml-1) using a marker DNA ladder of 100 bp. The products were visualized with ultraviolet illumination and imaged with gel documentation system (Biored Gel Doc XR, USA). Details of primer gene sequences and the different reaction temperatures carried out in the PCR assays used were as indicated in Table 1 [17].
| Primer | Nucleotide sequence | Target gene | Pathogenic E. coli strain | Denaturing | Annealing | Extension | Product size (Bp) | Cycles | Reference |
|---|---|---|---|---|---|---|---|---|---|
| EAE1 | F:5’AAACAGGTGAAACTGTTGCC3’ | eae eae | EPEC/EHEC | 94℃,2 min. | 55℃,60 s | 72℃,60 s | 490 | 35 | Khan, et al. |
| EAE2 | R:5’-CTCTGCAGATTAACCTCTGC-3’ | ||||||||
| EVS1 | F:5’-ATCAGTCGTCACTCACTGGT-3’ | Stx2 Stx2 | STEC/EHEC | 94℃,2 min. | 55℃,6℃ | 72℃,60 s | 110 | 30 | Khan, et al. |
| EVC2 | R:5’-CTGCTGTCACAGTGACAAA-3’ | ||||||||
| EHEC | F: 5’-ACGATGTGGTTTATTCTGGA-3’ | hlyA | EHEC | 95℃,3 min. | 45℃,40 s | 72℃,30 s | 165 | 30 | Paton and Paton, et al. |
| EHEC | R:5’-CTTCACGTCACCATACATAT-3’ | hlyA | |||||||
| EAEC | F:5’CTGGCGAAAGACTGTATCTAT-3’ R:5’CAATGTATAGAAATCCGCTGTT-3’ | aatA | EAEC | 95℃,3 min. | 45℃,40 s | 72℃,30 s | 630 | 30 | Chattaway,et al. |
| EAEC | aatA | ||||||||
| BFP | F:5’AATGGTGCTTGCGCTTGCTGC-3 R:5’GCCGCTTTATCCAACCTGGTA-3’ | bfpA bfpA | EPEC | 95℃,3 min. | 57℃,40 s | 72℃,30 s | 324 | 30 | Christian, et al. |
Note: EPEC: Enteropathogenic E. coli; EHEC: Enterohemorrhagic E. coli; STEC: Shiga-like toxin producing E. coli; EAEC: Enteroaggregative E. coli
Table 1: Primer gene sequence and PCR conditions.
Questionnaire survey
Questionnaire for data collection was prepared and administered to the attendants of children from whom specimens were collected. Data were collected on child demographics and clinical condition such as child age, sex, residence, onset of diarrhea, clinical diagnosis, history of previous illness, RVI status, BMI, household members and history of illness, etc.
Data management and analysis
Data describing the diarrheagenic conditions suggestive of E. coli infection observed on children along with age were classified filtered and coded using Microsoft Excel® 2007. The data were then exported to SPSS windows version 20.0 (IBMSPSS INC.Chicago, IL) for statistical analysis. The prevalence of E. coli strains from the total diarrheic children and the prevalence of pathogenic strains from total E. coli isolates along with their susceptibility profiles were determined by using descriptive statistics.
Children (n=98) (47 males and 51 females), aged 01 month to 60 months with a mean age of 31.9 months suffering from diarrhea were included in this study.
Occurrence of E. coli in diarrheic children
Out of 98 stool specimens collected from diarrheic children less than 5 years of age, 75 were found to be positive for E. coli based on colony characteristic on EMB; further confirmation by IMViC tests showed that only 56 of the isolates were E. coli.
The frequency of E. coli isolates in male children constituted 29.6% of diarrheic children included in the study while 27.5% of the isolates were found in female children. Table 2 shows the occurrence of E. coli isolates between male and female children and among three age groups of children involved in the study [19].
| Age in months | Sex | Total | |
|---|---|---|---|
| M | F | ||
| 0 month-6 months | 4 (4.1%) | 2 (2%) | 6 (6.1%) |
| 7 months-24 months | 13 (13.26%) | 10 (10.24%) | 23 (23.5%) |
| 25 months-60 months | 12 (12.2%) | 15 (15.3%) | 27 (27.5%) |
| Total | 29/98 (29.6%) | 27/98 (27.5%) | 56/98 (57.1%) |
Table 2: Age and sex distribution of diarrheic children with E. coli at AAU CHS Tikur Anbessa specialized hospital, Addis Ababa, 2017.
In addition, data on the clinical characters of the children were collected using questionnaires and analyzed. Table 3 shows the occurrence of E. coli among potential risk factors that could expose children to E. coli infection.
| Risk factors | E. coli occurrence | |
|---|---|---|
| Animal contact | YES 28 | 17 |
| NO 70 | 39 | |
| Habit of eating undercooked food | YES 33 | 20 |
| NO 65 | 36 | |
| Habit of boiling water | YES 16 | 9 |
| NO 82 | 47 | |
| Family history of illness | YES 27 | 12 |
| NO 71 | 44 | |
| Previous history of illness | YES 32 | 20 |
| NO 66 | 36 | |
Table 3: Clinical characters of diarrheic children and occurrence of E. coli at AAU CHS Tikur Anbessa general specialized hospital, Addis Ababa, 2017.
Antimicrobial susceptibility profiles of E. coli isolated from diarrheic children
E. coli isolates were tested against 11 antibiotics to determine their susceptibility patterns. The isolates were resistant to most of the antibiotics used. High percentage of resistance was observed for neomycin (94.6%), ampicillin (87.5%), and compound sulfonamides (83.9%).
The isolates showed low resistance towards ciprofloxacin and norfloxacin (33.9%, 26.8%), and to chloramphenicol (32.1%). Least resistance was exhibited by the isolates towards cefotetan among the antibiotics tested as indicated in Table 4 [20].
| Antibiotics | Susceptible | Resistant | Intermediate |
|---|---|---|---|
| Trimethoprim | 11/56 (19.6%) | 43/56 (76.8%) | 2 /56(3.6%) |
| Ciprofloxacin | 35/56 (62.5%) | 19/56 (33.9%) | 2/56 (3.6%) |
| Ampicillin | 4/56 (7.1%) | 49/56 (87.5%) | 3/56 (5.4%) |
| Neomycin | 1/56 (1.8%) | 53/56 (94.6%) | 2/56 (3.6%) |
| Gentamicin | 3/56 (5.4%) | 40/56 (71.4%) | 13/56 (23.2%) |
| Compound sulphonamide | 1/56 (1.8%) | 47/56 (83.9%) | 8/56 (14.3%) |
| Tetracycline | 8/56 (14.3%) | 48/56 (85.7%) | - |
| Chloramphenicol | 38/56 (67.9%) | 18/56 (32.1%) | - |
| Cefotetan | 46/56 (82.1%) | 10/56 (17.9%) | - |
| Norfloxacin | 40/56 (71.4%) | 15/56 (26.8%) | 1/56 (1.8%) |
| Streptomycin | 6/56 (10.71%) | 43/56 (76.8%) | 7/56 (12.5%) |
Table 4: Antibiotics susceptibility profiles of E. coli isolated from diarrheic children less than 5 years of age at AAU CHS Tikur Anbessa general specialized hospital, 2017.
Resistance pattern of E. coli isolates
Multidrug resistance was observed in all isolates of E. coli. All isolates were found to be resistant to at least 4 antibiotics. The highest prevalence of multi drug resistance was resistance to 7 antibiotics tested: Trimethoprim, ampicillin, neomycin, gentamycin, compound sulfonamide, streptomycin, and tetracycline among the antibiotics tested. One of the isolates exhibited resistance to all 11 antibiotics tested and 4 of the isolates exhibited resistance to 10 out 11 antibiotics tested. Data on the multidrug resistance pattern of the isolates are summarized in Table 5.
| Number of drugs resisted | Resisted drugs | Number of isolates that showed MDR | Percent |
|---|---|---|---|
| 4 | Neomycin, compound sulphonamide, gentamicin, ampicillin | 4 | 7.1 |
| 6 | Neomycin, compound sulphonaamide, gentamicin, ampicillin, streptomycin, tetracycline | 4 | 7.1 |
| 7 | Neomycin compund sulphonaamide, gentamicin, ampicillin, streptomycin, tetracycline, trimethoprim | 15 | 26.8 |
| 8 | Neomycin, compound sulphonaamide, gentamicin, ampicillin, streptomycin tetracycline, trimethoprim, ciprofloxacin | 14 | 25 |
| 11 | Neomycin, compound sulphonaamide, gentamicin, ampicillin, streptomycin tetracycline, trimethoprim, ciprofloxacin, chloramphenicol, norfloxacin, cefotitan | 1 | 1.8 |
| Total | 56 | 100 |
Table 5: Multi drug resistance of E. coli isolated from diarrheic children less than 5 years of age at AAUCHS Tikur Anbessa general specialized hospital, 2017.
Occurrence of E. coli pathotypes (pathogenic strains) in diarrheic children
Five pairs of primers (reverse and forward) were optimized according to their annealing temperatures and different virulence genes of E. coli were detected by PCR. Specimens were pooled to obtain control strains with the desired genes; in addition, positive controls from previous experiments at biotechnology laboratory, college of natural sciences were used in parallel with the current samples.
Many PCR experiments were run to detect virulence genes and to identify pathotypes of E. coli. It was difficult to incorporate all the images generated from the PCR experiments in the paper due to space limitation. Selected agarose gel images which represent each virulence gene generated from the different PCR runs according to specific base pairs are presented in Figures 1-5.
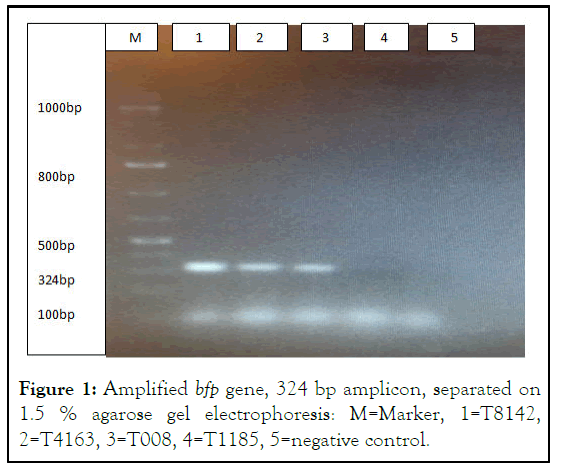
Figure 1: Amplified bfp gene, 324 bp amplicon, separated on 1.5 % agarose gel electrophoresis: M=Marker, 1=T8142, 2=T4163, 3=T008, 4=T1185, 5=negative control.
T8142 was obtained from pooled specimens during optimization process and was treated as a positive control in this run. T4163 and T008 are positive samples, T1185 is a negative sample.
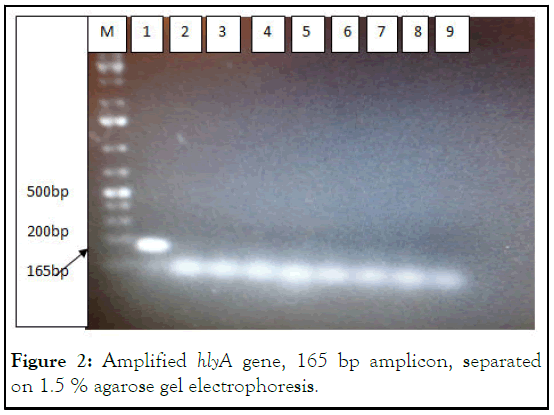
Figure 2: Amplified hlyA gene, 165 bp amplicon, separated on 1.5 % agarose gel electrophoresis.
M=Marker, 1=T004, 2=T4163, 3=T008, 4=T001, 5=T1185, 6=T002, 7=T5677, 8=T8142, 9=negative control. Only one isolate of E. coli, T004 was positive for hlyA gene out of 56 isolates.
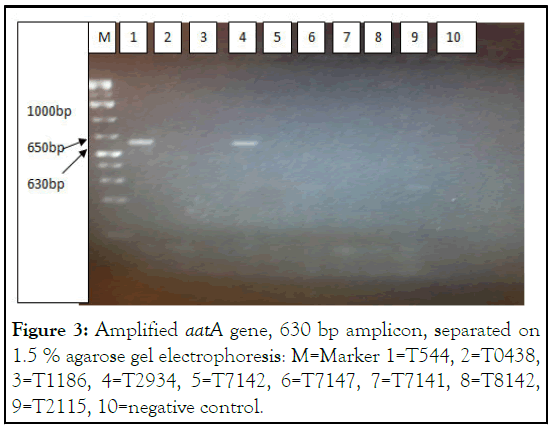
Figure 3: Amplified aatA gene, 630 bp amplicon, separated on 1.5 % agarose gel electrophoresis: M=Marker 1=T544, 2=T0438, 3=T1186, 4=T2934, 5=T7142, 6=T7147, 7=T7141, 8=T8142, 9=T2115, 10=negative control.
T544 was obtained from pooled specimens during optimization process and was treated as a positive control in this run. T2934 is also positive for aatA gene.
Figure 4: Amplified eae gene, 490 bp amplicon, separated on 1.5% agarose gel electrophoresis.
M=Marker, 1=T001, 2=T6028, 3=T7898, 4=T8316, 5=T0342, 6=T7849, 7=T0438, 8=T8311, 9=negative control. T001 was obtained from pooled specimens during optimization process and was treated as a positive control in this run. T0342 and T8311 are also positive for eae gene.
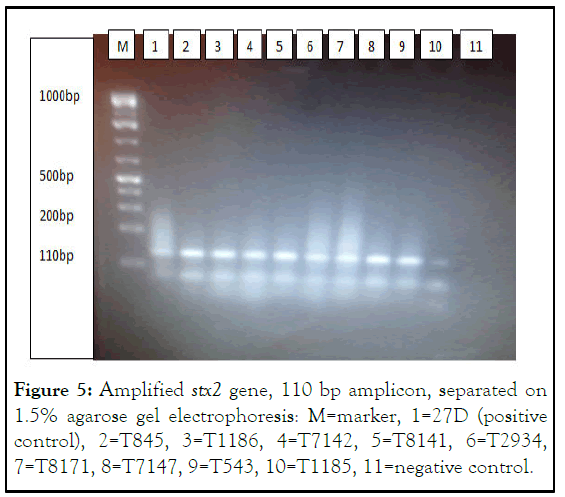
Figure 5: Amplified stx2 gene, 110 bp amplicon, separated on 1.5% agarose gel electrophoresis: M=marker, 1=27D (positive control), 2=T845, 3=T1186, 4=T7142, 5=T8141, 6=T2934, 7=T8171, 8=T7147, 9=T543, 10=T1185, 11=negative control.
Based on the different virulence genes detected, E. coli pathotypes/strains were identified as follows. Enteropathogenic E. coli (EPEC) strains were identified as those positive for eae (intimin) gene; shigatoxin producing E. coli (STEC) strains were identified as those positive for stx2; enterohemorrhagic E. coli strains were identified as those isolates positive for eae and stx2. Enteropathogenic E. coli are further classified into typical (positive for additional bundle forming pillus, bfp, gene) and atypical (negative for bfp) strains. Entero Hemorrhagic E. coli (EHEC) strains were further supported by their plasmid gene hemolysin (hlyA) gene. Isolates of E. coli with the plasmid gene aatA were considered as Entero Aggregative E. coli (EAEC). Finally isolates with more than one virulence gene were considered as mixed pathotypes. Five virulence genes were detected and 25 pathotypes (six categories) of E. coli were identified based on which virulence genes/s they contained; Entero Pathogenic E. coli (EPEC) was the most prevalent category with atypical EPEC being more common than typical EPEC. Mixed pathotypes with more than one virulence gene constituted for only 1% in the present study. Summary of the findings is presented in Table 6.
| Virulence gene detected | Frequency among E. coli isolates; N (%) | Frequency among diarrheic children; N (%) | Pathotypes/strains designation |
|---|---|---|---|
| Eae | 9 (16%) | 9/98 (9.1%) | Atypical EPEC |
| Eae+bfp | 3 (5.4%) | 3/98 (3.1%) | Typical EPEC |
| Stx2 | 8 (14.3%) | 8/98 (8.1%) | STEC |
| aatA | 3 (5.4%) | 3/98 (3.1%) | EAEC |
| Stx1+eae | 1 (1.8%) | 1/98 (1%) | EHEC |
| hlyA | 1 (1.8%) | 1/98 (1%) | EHEC |
| Stx1+aatA | 1 (1.8%) | 1/98 (1%) | Mixed pathotypes |
| Total pathotypes | 25/56 (44.6%) | 25/98 (25.5%) |
Table 6: Frequency of different E. coli pathotypes detected from diarrheic children less than 5 years of age at AAU CHS Tikur Anbessa general specialized hospital, 2017.
The distribution of E. coli pathotypes among the age groups of children is shown in Table 7.
| Age group | Pathotypes detected | Total | |||||
|---|---|---|---|---|---|---|---|
| Atypical EPEC (eae+) | STEC (stx2+) | Typical EPEC (eae+, bfp+) | EAEC (aatA+) | Mixed pathotypes (aatA+, stx1+) | EHEC (hlyA+, stx) | ||
| 0 months-6 months | 1 | 0 | 0 | 1 | 0 | 0 | 2 |
| 7 months-24 months | 4 | 3 | 1 | 2 | 1 | 1 | 12 |
| 24 months-60 months | 4 | 5 | 2 | 0 | 0 | 0 | 11 |
| Total | 9 (36%) | 8 (32%) | 3 (12%) | 3 (12%) | 1 (4%) | 1 (4%) | 25 (100%) |
Table 7: Occurrence of E. coli pathotypes among different age categories of diarrheic children less than 5 years of age at AAU CHS Tikur Anbessa general specialized hospital, 2017.
Detection of the virulence gene eae was also analyzed among the clinical characters of the children and the results are presented in Table 8.
| Risk factor | eae occurrence | |
|---|---|---|
| Animal contact | YES 17 | 4/56 |
| NO 39 | 8/56 | |
| Habit of eating undercooked food | YES 20 | 5/56 |
| NO 36 | 7/56 | |
| Habit of boiling water | YES 9 | 2/56 |
| NO 47 | 10/56 | |
| Family history of illness | YES 12 | 3/56 |
| NO 44 | 9/56 | |
| Previous history of illness | YES 20 | 5/56 |
| NO 36 | 7/56 | |
Table 8: Clinical characters of diarrheic children less than 5 years of age and occurrence of E. coli eae gene at AAU CHS Tikur Anbessa general specialized hospital, 2017.
Antibiotic susceptibility profiles of E. coli pathotypes identified from diarrheic children
Drug susceptibility profiles of the different pathotypes detected showed that the pathotypes exhibited higher resistance to most of the antibiotics tested. Lower resistance was exhibited by the pathotypes towards Cefotetan. One interesting observation is to see that plasmid encoded virulence genes, bfp and aatA, are associated with increase in resistance, actually 100% resistance which may be a potential indicator for co-carriage of both virulence and resistance gene on the plasmid. The results are summarized in the Table 9.
| Antibiotic resistance frequency of E. coli pathotypes | ||||
|---|---|---|---|---|
| Antibiotics | Typical EPEC (eae+) | STEC (stx1+) | Atypiccal EPEC (eae+ fp+) | EAEC (aatA+) |
| Trimethoprim | 9 (75%) | 7 (77.8%) | 3 (100%) | 2 (50%) |
| Ciprofloxacin | 6 (50%) | 5 (55.6%) | 1 (33.3%) | 4 (100%) |
| Ampicillin | 11(91.7%) | 6 (66.7%) | 3 (100%) | 4 (100%) |
| Neomycin | 12 (100%) | 8 (88.9) | 3 (100%) | 2 (50%) |
| Gentamycin | 10 (83.3%) | 5 (55.6%) | 3 (100%) | 4 (100%) |
| Compound sulphonamide | 10 (83.3%) | 8 (88.9) | 3 (100%) | 4 (100%) |
| Tetracycline | 11(91.7%) | 8 (88.9) | 3 (100%) | 4 (100%) |
| Chloramphenicol | 7 (58.3%) | 3 (33.3%) | 1 (33.3%) | 3 (75%) |
| Cefotitan | 2 (16.7%) | 2 (22.2%) | 1 (33.3%) | 1 (25%) |
| Norfloxacin | 6 (50%) | 2 (22.2%) | 1 (33.3%) | 4 (100%) |
| Streptomycin | 10 (83.3%) | 2 (22.2%) | 3 (100%) | 2 (50%) |
Table 9: Distribution of resistance profiles among different pathotypes of E. coli detected from children less than 5 years of age at AAU CHS Tikur Anbessa general specialized hospital, 2017.
Multi drug resistance was observed in all categories of E. coli pathotypes. All of the pathotypes were found to be resistant to at least 4 antibiotics.
The highest prevalence of multi drug resistance was resistance to 8 antibiotics tested: Trimethoprim, ampicillin, neomycin, gentamycin, compound sulfonamide, streptomycin, chloramphenicol and tetracycline among the antibiotics tested. One of the isolates exhibited resistance to all 11 antibiotics tested. The results are summarized in Table 10.
| Number of drugs resisted | Resisted drugs | Number of isolates that showed MDR | Percent |
|---|---|---|---|
| 4 | Neomycin, compound sulphonamide, gentamicin, ampicilin | 2 | 8 |
| 6 | Neomycin, compound sulphonamide, gentamicin, ampicillin streptomycin, tetracycline | 1 | 4 |
| 7 | Neomycin, compound sulphonamide, gentamicin, ampicillin streptomycin, tetracycline, trimethoprim | 6 | 24 |
| 8 | Neomycin, compound sulphonamide, gentamicin, ampicillin streptomycin, tetracycline, trimethoprim, ciprofloxacin | 9 | 36 |
| 11 | Neomycin, compounds sulphonamide, gentamicin, ampicillin streptomycin, tetracycline, trimethoprim, ciprofloxacin | 1 | 4 |
| Total | Chloramphenicol, norfloxacin, cefotitan | 25 | 100 |
Table 10: Multi drug resistance of E. coli pathotypes isolated from children less than 5 years of age at AAUCNS Tikur Anbessa specialized hospital, 2017.
Despite the significant contribution that pathogenic E. coli strains have to the burden of diarrhea, their distribution in Africa is not well studied. Studies undertaken in few countries like Kenya, Nigeria and South Africa have been concentrated on certain localities and specific risk populations. This has led to the lack of capacity to detect diarrheagenic E. coli in patients with diarrhea. Diarrheagenic E. coli is rarely included in the range of target organisms in many studies in Africa. Pathogenic E. coli is not usually considered as a possible cause of child diarrhea but diagnostic tests requested at Tikur Anbessa hospital are mostly for Salmonella and Shigella i.e., there is no appropriate laboratory diagnosis for pathogenic E. coli. In the present study, E. coli was found in 56/98 (57.1%) pediatric children less than 5 years of age at Addis Ababa university, college of health sciences, Tikur Anbessa specialized hospital. Lower results were reported by other studies in Ethiopia (204/422, 48.3%), Sudan (211/437, 48%) and Italy (75/160, 46.9%). The occurrence of E. coli was similarly rated in all age groups examined and in both sexes of the children involved in the current study. This finding is in contrast with another study conducted in Ethiopia which reported that the isolation rate of E. coli was high in children aged 6 months-23 months. Antibiotic resistance rate of E. coli isolates in this study was generally high (>40%). Similar results were reported by studies made in Kenya and Eastern Romania which indicated that E. coli isolates showed high level of resistance to commonly use and locally available antimicrobial agents. But this finding is in contrast with reports of a study conducted in Nicaragua which show that E. coli isolates exhibited low resistance to most antimicrobial drugs and that E. coli have not reached high level of resistance to commonly used antibiotics. In the present study, E. coli isolates showed high resistance rates towards many antimicrobial drugs including ampicillin, trimethoprim, gentamycin and tetracycline. This result is in agreement with a previous study made in Ethiopia in which high level of resistance to ampicillin (86.8%), and tetracycline (76%) was documented. But the finding is in contrast with reports of a study made in Nepal in which 91.7% E. coli isolates were susceptible to gentamycin and 75% isolates were susceptible to tetracycline. It is also in contrast with a study made in Sudan which reported that 94% E. coli isolates were susceptible to gentamicin. Low resistance was exhibited by E. coli isolates in the present study towards chloramphenicol, ciprofloxacin, and norfloxacin; cefotetan being the antibiotic to which the isolates showed the highest susceptibility. Resistance to ciprofloxacin and norfloxacin at low levels was documented in a similar study in Kenya.
On the other hand, 100% susceptibility of E. coli isolates to chloramphenicol was reported by studies from Nepal and Sudan in contrast to the present study. Multidrug resistance was observed in all isolates of E. coli in the present study. This is in contrast with a study conducted in Mexico in which multidrug resistance was reported in 58% of isolates. The highest prevalence of multi drug resistance of E. coli isolates in the present study was resistance to 7 antibiotics tested: trimethoprim, ampicillin, neomycin, gentamycin, compound sulfonamide, streptomycin, and tetracycline. This is in contrast with a study made in Nicaragua in 2011 in which the most prevalent multidrug resistance in E. coli isolates was resistance to 2 antibiotics. Factors like mutation which give selective advantage to resistant strains may be involved in the high antibiotic resistance observed in the present study. The high antibiotic resistance rate observed in the present study may indicate that the number of potential commensal reservoirs of resistance genes which can be transferred to pathogens is on increase and that an increase in the emergence of drug resistant pathogenic E. coli strains is a possibility. The finding in the present study may be a useful indicator of bacterial antibiotic resistance in the local community. 25/56 (44.6%) of the E. coli isolates in this study were found to be pathogenic classes carrying different virulence genes. This study investigated the identity of pathogenic E. coli strains that occurred in child diarrhea using gene specific primers. Thus, the pathogenic strains named STEC, EPEC, EHEC, and EAEC were identified from processed samples. Higher frequency of pathogenic strains than the present study was reported in a study conducted in Costarica in 2010 (77%). Lower figures were reported by studies from Tanzania (22.9%) and Burkina Faso (24%) in 2011 and 2013 respectively. A study conducted in Mexicoin 2005 reported 14% isolation rate of diarrheagenic E. coli from children and a similar study conducted in North Western Italy in 2011 reported 13.33% isolation rate. Atypical EPEC were the most prevalent pathotypes in the present study being found in 16% of E. coli isolates followed by STEC which constituted for 14.3% of E. coli isolates. Similar finding was reported in Costarica in 2010. But the finding in the present study is in contrast with findings from Burkina Faso and Sudan in 2013 and 2015 respectively which reported that EAEC were the most frequent pathotypes. A study in India also reported higher occurrence of typical EPEC strains than atypical ones in 2010 which is in contrast with the present study.
The high isolation rate of pathogenic strains in the present study may be attributed to increased evolution of pathogens through horizontal gene transfer of mobile genetic elements harboring virulence genes and may be an indication that there is tremendous environmental ecological niche which is a reservoir of virulence genes. Extensive hospital and environment based virulence studies are required to come up with more conclusive data. The isolation of mixed pathotypes (isolates possessing more than one virulence gene) was 1.8% in the present study (1/56). A 3.8% isolation rate of mixed pathotypes was reported in India in 2010. A study made in Brazil IN 2012 found that infection by mixed pathotypes was generally rare. Taking more than one colony of bacteria from the original culture for extraction of bacterial DNA may increase the likelihood of identifying mixed pathotypes from a single patient. Diarrheagenic E. coli in the present study exhibited high resistance rates towards many of the antimicrobials tested; low resistance rates were observed towards cefotitan. This is in agreement with results of a study made in Vietnam in 2005 in which low sensitivity of E. coli isolates was documented towards ampicillin, chloramphenicol and ciprofloxacin. A study made in Iran in 2011 reported 100% resistance of EPEC strains to ampicillin which is in agreement with the present study in which resistance to ampicillin was 100% for atypical EEC strains. Pathotypes with plasmid coded virulence genes showed increased resistance rates in the present study as indicated in Table 11 in the result section. This may be an indication of cocarriage of virulence and resistance genes on the plasmids of the pathotypes. Antibiotic resistance may be usually associated with environmentally acquired extra chromosomal mobile genetic elements including plasmids, transposons, and integrons apart from chromosomal mutations which are usually considered to be rare. E. coli are known to show high genetic flexibility in that resistance and virulence genes can be transferred together from one bacteria to another through horizontal gene transfer. Resistance occurs more effectively due to transfer of resistance genes than chromosomal mutation. Plasmids encoding genes that confer resistance to different classes of antimicrobial agents including cephalosporins, fluoroquinolones, aminoglycosides and virulence determinants that help bacterial cells to have adaptability and fitness in different ecologies can render bacteria both pathogenic and resistant. E. coli plasmids can thus carry both virulence and resistance genes. Accessory genetic materials in bacteria can acquire resistance genes there by promoting their transmission between bacteria. Among other mobile genetic elements, plasmid mediated transmission is the most common mechanism of such horizontal transfer of resistance genes. Coevolution of both virulence and resistance can be an explanation for the increased resistance pattern in the pathotypes with plasmid coded virulence genes aatA and bfp in the present study. This can be an area of extended research on association between virulence and resistance. Pathotypes generally showed high resistance patterns in the present study with atypical EPEC and EAEC showing the highest resistance rates. This is in contrast with reports of a study made in Costa Rica in 2010 in which pathogenic E. coli exhibited less prominent resistance. But the finding of the present study is in agreement with a finding from Peru in 2009 in which differential resistance patterns were exhibited by individual E. coli pathotypes with EAEC strains exhibiting higher resistance level than EPEC. The high antibiotic resistance rates in the present study may be attributed to the presence of commensal resistant bacteria which act as reservoirs of resistance genes that are transferred to pathogenic strains. 100 % multi drug resistance was observed in all pathotypes of E. coli in the present study. The most prevalent multidrug resistance among isolated pathogenic strains was resistance to 8 antibiotics tested: Trimethoprim, ampicillin, neomycin, gentamycin, compound sulfonamide, streptomycin, chloramphenicol and tetracycline among the antibiotics tested. This result is higher than results reported by other studies. 70.6% multi drug resistance was reported in a study made in Iran in 2011 in isolated EPEC pathotypes. Similarly 86.4% multidrug resistance of E. coli pathotypes was reported in studies made in Iran in 2014. Factors like duration of hospital stay, selfmedication, poor patient’s compliance; environmental conditions may be involved in the emergence of multi drug resistant strains. Pathotypes which harbor chromosomal virulence genes were identified in higher numbers in the present study while pathotypes which harbor plasmid virulence genes like EAEC were comparably few. This may be due to loss of integrity of plasmids as a result of delay in bacterial DNA extraction after isolation of the bacteria.
The isolation rate of E. coli in this study was high. E. coli exhibited high rates of antibiotic resistance to many of the antibiotics tested including ampicillin, gentamycin, chloramphenicol and tetracycline. Moreover, all E. coli exhibited multiple drug resistance. This is an indication that there is a need for extensive study on the occurrence, risk factors and genetic back ground of antimicrobial resistance of E. coli in the study area.
The involvement of antibiotic resistant pathogenic E. coli in diarrheic children is prominent and hence diagnosis and antimicrobial sensitivity testing procedures need to be incorporated in to routine laboratory practices at AAU CHS Tikur Anbessa general specialized hospital. Determination of antibiogram before antibiotic Prescription for effective treatment is recommended. Among antibiotics tested, cefotitan was found to be the most effective drug against isolates of E. coli.
Before the start of the work, the proposal was submitted to the Ethics committee of Addis Ababa University College of Natural Sciences to get Ethical approval to conduct the study (CNSDO/ 237/09/2017). During sample collection the objectives of the work was explained to the parents of children visiting hospitals in order to get consent of the parents or attendants of children. In addition, all samples were collected by health professionals.
I, Benyam Zenebe, declare that the information in this manuscript is true and correct. I believe it contains no material I would not want to include in my research publication. I acknowledge that this is a result my MSc thesis work which was conducted under the supervision of my advisers professor Tesfaye Sisay, Professor Gurja Belay, and Professor Workabeba Taye.
Before the start of the work, the proposal was submitted to the Ethics committee of Addis Ababa University College of natural sciences to get ethical approval to conduct the study (CNSDO/ 237/09/2017). During sample collection the objectives of the work was explained to the parents of children visiting hospitals in order to get consent of the parents or attendants of children. In addition, all samples were collected by health professionals.
The datasets used of this study are available from the corresponding author on reasonable request.
The authors declare that they have no competing interests.
This study was funded by the thematic research project entitled “biotechnological approaches to harness microbe host environment interactions for sustainable development of health and agricultural productivity” at the institute of biotechnology, college of natural and computational sciences, Addis Ababa University; the study was also supported by the department of microbial, cellular and molecular biology, college of natural and computational sciences, Addis Ababa university.
The authors would like to acknowledge Addis Ababa college of natural sciences microbial, cellular and molecular biology, Addis Ababa university college of natural sciences institute of biotechnology; Addis Ababa university college of health sciences Tikur Anbessa specialized hospital.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Zenebe B, Sisay T, Belay G, Abebe W (2024) Pathogenic Escherichia coli Strains and their Antibiotic Susceptibility Profiles in Cases of Child Diarrhea at Addis Ababa University, College of Health Sciences, Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. Clin Pediatr. 9:271.
Received: 04-Jul-2023, Manuscript No. CPOA-23-25401; Editor assigned: 06-Jul-2023, Pre QC No. CPOA-23-25401 (PQ); Reviewed: 20-Jul-2023, QC No. CPOA-23-25401; Revised: 11-Jul-2024, Manuscript No. CPOA-23-25401 (R); Published: 18-Jul-2024 , DOI: 10.35248/2572-0775.24.9.271
Copyright: © 2024 Zenebe B, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.