Virology & Mycology
Open Access
ISSN: 2161-0517
ISSN: 2161-0517
Research Article - (2022)Volume 11, Issue 6
The opportunistic fungus Pneumocystis jirovecii causes PneumocystisPneumonia (PCP), a life-threatening infection in HIV/AIDS and other immunocompromised patients. The seemingly low prevalence of P. jirovecii pneumonia in sub-Saharan Africa has been a matter of great debate; unsuitable diagnostic practices have been added to the uncertainty of PCP prevalence. Few studies have evaluated the utility of easily obtainable samples such as expectorated sputum for diagnosis. Smear -ve pulmonary TB is diagnosed according to clinical and radiographic criteria, due to unavailability of mycobacterial cultures. In high seroprevalence areas of HIV/AIDS there’s a concern that other opportunistic infections apart from TB, such as P. jirovecii may be missed. We proposed to determine the prevalence of P. jirovecii in confirmed TB smear negative and retreatment cases at the CGH. Sputum Samples were analyzed by microscopy after Toluidine Blue O staining and by PCR to evaluate the techniques in detecting P. jirovecii. 200 TB smear negative samples were analyzed irrespective of the patients HIV status. Sample analysis by both the staining technique and PCR were compared. We aimed to determine the significance of PCP in TB smear negative and retreatment cases and significance of P. jirovecii in TB smear negative and retreatment cases at the CGH.
Pneumocystis jirovecii; Nested PCR; Pulmonary tuberculosis; Smear negative
Genus, Pneumonia caused by Pneumocystis jirovecii (PCP) (previously known as P. carinii) has long been recognized in patients with impaired immunity. It was initially described as a cause of epidemic interstitial pneumonia in premature and malnourished infants. Until 1980, PCP was uncommon and recognized in patients who were immune compromised because of malignancies, immunosuppressive therapy, or congenital immunodeficiency. However, the rate of infection by P. jirovecii increased with the emergence of the Human Immunodeficiency Virus (HIV) [1].
In developed countries, the epidemiology and clinical spectrum of PCP have been clearly defined and well documented. In contrast, a limited number of epidemiological studies have evaluated PCP prevalence in developing countries [2].
Recent reports have described an increased rate of PCP in Africa, Asia and South America [3-6].
In Tuberculosis (TB) endemic parts of the world, substantial numbers of patients with pulmonary symptoms are managed as “smear negative TB patients” or treated for TB without a definitive diagnostic criterion [7-9]. Lack of access to diagnose Pneumocystis jirovecii pneumonia might be a contributing factor. We therefore propose to determine the prevalence of P. jirovecii by PCR in sputum of TB smear negative or retreatment patient at the Coast General Hospital.
Study design
This was a cross sectional laboratory study nested under a main approved study KEMRI/SERU/CMR/0037/3213. Patients were sampled at the TB clinic during diagnosis. The study was done at the Mycology laboratory, Center for Microbiology Research KEMRI.
Study area
The study was carried out at the Coast General Hospital as it’s the main epicenter of most patients with TB referral cases in the region. It also captures a large population of the Coast and Mombasa County.
Study population
The study population was the patients confirmed to be TB smear negative but have or suspected to have pulmonary tuberculosis infection. All patients that had been diagnosed as smear negative TB, who provided sputum specimens for diagnosis and those on retreatment cases attending the TB clinic at the Coast General Hospital. The patients included all adults above 18 years that gave consent and provided sputum sample.
Sample collection
A total of 200 samples were collected, two specimens from each patient. A specimen was a deep cough specimen. The patients were instructed to brush and rinse mouths thoroughly, inhale and exhale deeply forcing air from the lungs using the diaphragm.
This was repeated until the patient coughs and able to produce a sputum specimen. At least 2 mls of sputum was collected within 15 to 30 minutes after waking and before taking breakfast. Saliva and any specimens that are of no diagnostic value were discarded. The qualities of the sputum specimens were quantitatively accessed based on the number of squamous epithelial cells and any salivary specimens were discarded. The samples were labeled with patient number.
Microbiological investigations
Toluidine blue O staining: The conventional staining methods and the PCR were compared in terms of sensitivity and specificity, with TBO staining which is used as the gold standard method for diagnosis of pneumocystosis [10-12].
Sputum smears was made on glass slides and heat fixed and then allowed to air dry. Smears were placed in sulfating reagent made by mixing 45 ml of glacial acetic acid with 15 ml of concentrated sulfuric acid for 10 minutes then rinsed in cold water for 5 minutes and then drained the excess water. Smears were placed in Toluidine Blue (0.3 g of dye in 60 ml of water) for 3 minutes and rinsed with 95% ethanol followed by absolute ethanol and then xylene. The smears were air dried and examined at × 40 and × 100 for the presence of Pneumocystis jirovecii.
A positive confirmation was taken if at least one cluster with characteristic cyst was detected and defined as equal to or ≥ 2.The smears were examined for presence of oocysts and the slides with P. jirovecii scored as positive, if positive the number of clusters were counted and graded.
DNA preparation and extraction: Fungal DNA was extracted from the samples using the MycXtra fungal DNA extraction kit (Myconostica Ltd., Manchester, United Kingdom) according to the manufacturer’s instructions.
PCR methodology
Sputum samples were subjected to DNA extraction and isolation using a commercial Genomic DNA Kit in accordance with the manufacturer’s instructions. PCR was performed based on amplification of a portion of the Large Sub Unit (LSU) of the mitochondrial ribosomal RNA gene of P. jirovecii. Primers against different genes have been used by various authors in the past. These are mtLSUrRNA, Thymidylate Synthetase (TS), 18SrRNA, 5S rRNA, Major Surface Glycoprotein (MSG) and Internal Transcribed Spacer (ITS) [13]. Out of these genes, PCR for mt LSU has shown most promising results and the same was used for this study.
The primers and beacons avoid 5 P. jirovecii polymorphisms in the mtLSUrRNA and are not affected by P. jirovecii heterogeneity while avoiding amplification of human DNA.
Denaturing, annealing and extension times were run at 1 min each, at 95, 58 and 72˚C, respectively. DNA samples were amplified for 40 cycles. For nested PCR were repeated the latter conditions for 35 cycles using the product of the 1st PCR round as the template. The resulting amplified product of 364 bp fragment was separated on a run on 3% electrophoresis on gel, and was assessed in the Trans illuminator device. Each run of PCR was performed with a positive and negative control so as to check performance of the test.
Primers: PCR was performed according to Wakefield et al., [14]; the Genome amplification was done using PCR primers pAZ102E and pAZ102H, derived from the mitochondrion large subunit rRNA (mtLSUrRNA) gene (Table 1).
| Primer | Primer sequences (5’-3’) |
|---|---|
| pAZ102-E | GATGGCTGTTTCCAAGCCCA |
| pAZ102-H | GTGTACGTTGCAAAGTACTC |
Table 1: List of PCR primers.
Negative and positive controls
In the present study, the positive control was a sample of which P. jirovecii has been isolated or stocked isolate. Deionized distilled water was used instead of a template as the negative control.
Statistical analysis
Data analysis plan: Statistical assessment of these characteristics will be performed using the _2 test (P, 0.05 will be considered significant). Statistical assessment of differences of the mean of Pneumocystis DNA concentration [log (copies) per capillary] from PCP or non-PCP cases will be performed by unpaired Student’s t-test (P, 0.05 was considered significant).
The sensitivity, specificity, positive and negative predictive values for TBO and PCR will be calculated with 95% Confidence Intervals (CI). The statistical concordance between clinical diagnosis and different techniques will be analyzed by a kappa coefficient of Cohen (95% CI).
Demographic parameters of the patients
A total of 350 participants were enrolled to the study; 100 participants were able to expectorate sputum sample. Median ages was 38 years old, (IQR: 18-78) of whom 62% 62 were males and 38% 38 were females. Most of the patents were labelled as TB smear negative or and retreatment cases.
The results of this study showed that patients with pulmonary tuberculosis were co-infected with fungi; P. jirovecii as opportunistic fungal organisms.
A total of 62 males (62%) and a total of females 38 (38%) participated in the study which involved a total of 100 samples. Males appeared to be more susceptible to TB a high prevalence which could have been attributed to their health seeking behaviors.
A total of 29 samples (11.0%) were positive for Pneumocystis jirovecii. Toluidine blue stain detected P. jirovecii cysts in 27.17% of the patients. The Nested PCR detected 38% positive for P. jirovecii. The sensitivity and specificity of nested PCR as compared to TBO staining was 4.27% and 20.6% respectively. Nested PCR test detected additional 12 more patients than toluidine O staining technique showing high sensitivity, TBO had an acceptable sensitivity and very high specificity.
Microbiological results
Toluidine blue 0 stains also apparently stained only the cyst forms. Photographs of P. jirovecii stained by Toluidine blue O Stain Results with the TBO are shown in Figures 1-3.
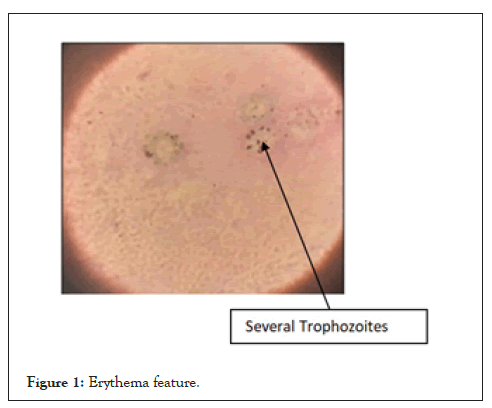
Figure 1: Erythema feature.
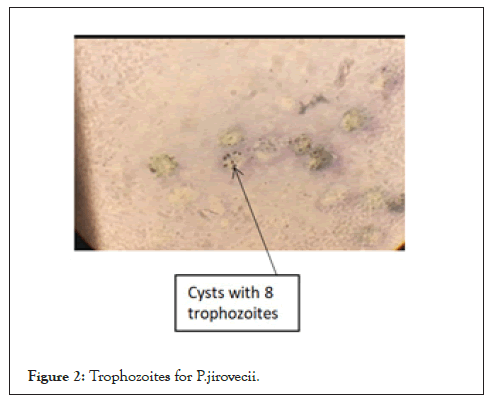
Figure 2: Trophozoites for P.jirovecii.
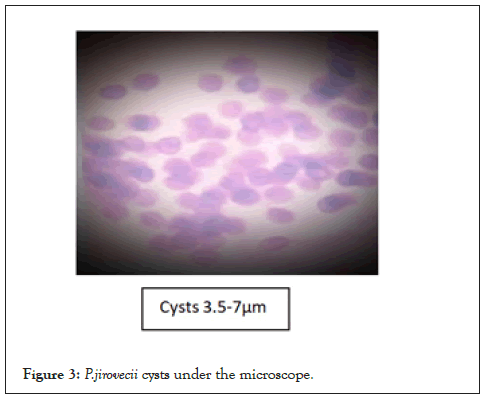
Figure 3: P.jirovecii cysts under the microscope.
In Figures 1 and 2, the cysts are lavender in color but are somewhat grainy in appearance; much background material is also evident. In Figure 3, structures thought to be P. carinii cysts have a spherical appearance and the outlines of individual cysts are quite faint. In Figure 3 the cyst forms appear as lavender structures approximately 5 µm in diameter, and many are cup-shaped. The cyst outline is distinct, and the internal region stains uniformly (Figures 1-3).
The cysts were frequently observed in clusters, and they did not bud. Very little background material was seen with this stain; particularly with touch the sulfation reagent which digests away most of the background material in both lavages and lung touch preparations but does not affect either fungal elements or P. jirovecii. The toluidine blue O stains the residual material remaining on the slide. Most of the positive slides appeared as shown below:
Molecular results
The primers described before on Table 2 lists of primers were used to amplify a 364 bp segment of a specific region at the Mitochondrial Large subunit gene of P. jirovecii. Amplification was done by adding 1 µl extracted DNA, 21 µl of nuclease free water, 0.5 µl of forward primer And 0.5 µl of reverse primer into a master mix which were lyophilized with 20 µl reaction volume containing a reaction buffer, MgCl2, DNTPs and a DNA polymerase. This mixture made a total volume of 25 µl. The 2 µl was for pipetting errors. Nested PCR was performed in the first round of amplification. Denaturing, annealing and extension times were run at 1 min each, at 95, 58 and 72˚C, respectively for 40 cycles and repeated the latter conditions for 35 cycles using the product from the 1st PCR round as the template using the same primers as the first round (Table 2).
| ALL | |
| N (%) | 100(100%) |
| Males | 62(62%) |
| Females | 38(38%) |
| TB smear negatives | 28(%) |
Table 2: Demographics of the study participant.
The first gel Fig 5 was for the positive microscopic slides that clearly had the cysts and several trophozoites. The separation on gel agarose was done for 45 minutes and the gel was prestained with ethidium bromide. The 2nd gel in Figure 4, where the above conditions were followed for the conditions and the gels prestained with ethidium bromide and viewed under the Trans illuminator. In all the gel runs the 100 bp ladder was used followed by the negative control and a positive control. A total of 29 samples out of the total 100 expressed the 364 band fragment on the agarose gel (Figure 4).
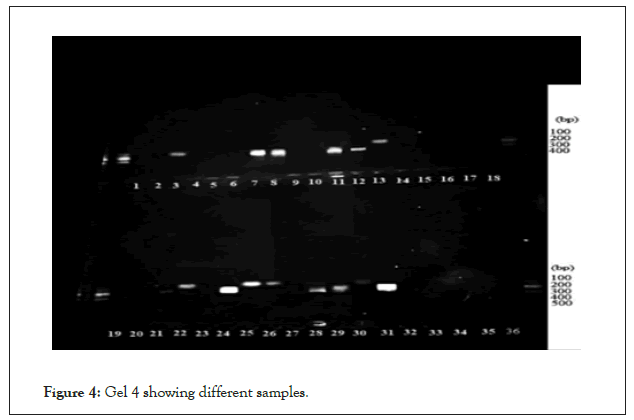
Figure 4: Gel 4 showing different samples.
Hollyhock Toluidine blue 0 stains also apparently stained only the cyst forms. Results with the TBO are shown in Figures 1-3.
In Figures 1 and 2, the cysts are lavender in color but are somewhat grainy in appearance; much background material is also evident. In Figure 3, structures thought to be P. carinii cysts have a spherical appearance and the outlines of individual cysts are quite faint. In Figures 3 and 4 the cyst forms appear as lavender structures approximately 5 µm in diameter.
Pneumocystis organisms have a high tropism for the lung. All the stages of the lifecycle of the organism have been observed within the lungs and exist exclusively within the lungs of infected hosts. The organism resides extracellularly, predominantly in the lung alveoli.
The life cycle of Pneumocystis mainly consists of 3 major life cycle stages identified by morphology; the cyst, the pre-cystic and the trophic form stages. The trophic form is mono nuclear and thin walled and usually dominates during active infection and fills the alveoli space and is variable in length 1 to 8 µm long with multiple pseudo hyphal structures which are used as an anchor to host cells.
The alcohol and xylene washing procedures remove excess toluidine blue O from the slide. In some other cases, P. jirovecii was difficult to detect because of the presence of background debris.
P. carinii cysts typically occur both singly and in small clusters of closely associated organisms. When only single cysts are seen and in specimens containing yeast cells, great care must be exercised in making a diagnosis of P. jirovecii. In specimens containing yeast cells, we think that a positive diagnosis of P. jirovecii cannot be made without finding at least one cluster of the characteristic cysts of P. jirovecii [15-18].
Ultra structural analysis of the trophic form shows a single nucleus surrounded by a thin nuclear membrane, rough and smooth endoplasmic reticulum, glycogen granules and usually a single mitochondrion [19]. The majority of trophic forms observed in an infected organism are haploid, but some are diploid [20]. It is not certain whether the diploid trophic organisms are the result of mating between two trophic forms or represents a step towards asexual division of a single organism; however, there is increasing evidence that Pneumocystis does have a sexual life cycle phase.
The next phase in the life cycle after the trophic form is the precyst, which may be an intermediate stage in sexual reproduction. The precyst is larger than the trophozoites, ∼4–8 µm, and more spherical in shape [21]. A mature cyst typically contains eight intracystic bodies within its cell wall, but can also contain smaller numbers such as two or four spores [21].
The primers described before on Table 3 list of primers were used to amplify a 364 bp segment of a specific region at the Mitochondrial Large subunit gene of P. jirovecii. Amplification was done by adding 1 µl extracted DNA, 21 µl of nuclease free water, 0.5 µl of forward primer And 0.5 µl of reverse primer into a master mix which were lyophilized with 20 µl reaction volume containing a reaction buffer, MgCl2, DNTPs and a DNA polymerase. This mixture made a total volume of 25 µl. The 2 µl was for pipetting errors. Nested PCR was performed in the first round of amplification. Denaturing, annealing and extension times were run at 1 min each, at 95, 58 and 72˚C, respectively for 40 cycles and repeated the latter conditions for 35 cycles using the product from the 1st PCR round as the template using the same primers as the first round (Table 3).
| Demographics | Sputum characteristics | TB | Pneumocystis detection | ||||
|---|---|---|---|---|---|---|---|
| Patient number | Age | Sex | Specimen volume (ml) | Specimen appearance | AFB smear | Nested PCR | Toluidine Blue O microscopy |
| Result | |||||||
| 1 | 32 | F | 3.5 | Mucoid | N | N | P |
| 2 | 36 | F | 2 | Muco-salivary | N | P | N |
| 3 | 45 | F | 2 | Muco-salivary | N | P | N |
| 4 | 44 | F | 2 | Muco-salivary | N | P | N |
| 5 | 59 | M | 2 | Muco-salivary | N | P | N |
| 6 | 50 | M | 2 | Muco-salivary | N | P | N |
| 7 | 36 | M | 2 | Muco-salivary | N | N | P |
| 8 | 57 | M | 7.5 | Mucoid | N | P | N |
| 9 | 33 | M | 2 | Muco-salivary | N | P | N |
| 10 | 39 | F | 6.5 | Muco-salivary | N | P | N |
| 11 | 29 | M | 2 | Mucoid | N | N | P |
| 12 | 73 | M | 7 | Muco-salivary | N | N | P |
| 13 | 20 | F | 2 | Muco-salivary | N | P | N |
| 14 | 31 | M | 2 | Mucoid | N | P | N |
| 15 | 18 | F | 4 | Muco-salivary | N | N | P |
| 16 | 28 | M | 2 | Muco-salivary | N | P | N |
| 17 | 70 | M | 5 | Muco-salivary | N | P | N |
| 18 | 49 | F | 2 | Muco-salivary | N | N | P |
| 19 | 35 | F | 2 | Mucoid | N | N | P |
| 20 | 46 | F | 2 | Muco-salivary | N | P | N |
| 21 | 48 | F | 4.5 | Mucoid | N | P | N |
| 22 | 27 | M | 4 | Muco-salivary | N | N | P |
| 23 | 64 | F | 3.5 | Mucoid | N | P | N |
| 24 | 30 | M | 2 | Muco-salivary | N | P | N |
| 25 | 48 | F | 2 | Mucoid | N | P | N |
Note: P=Positive; N=Negative.
Table 3: Characteristics of samples that tested positive for PCP.
The first gel Figure 5 was for the positive microscopic slides that clearly had the cysts and several trophozoites. The separation on gel agarose was done for 45 minutes and the gel was prestained with ethidium bromide (Figure 5).
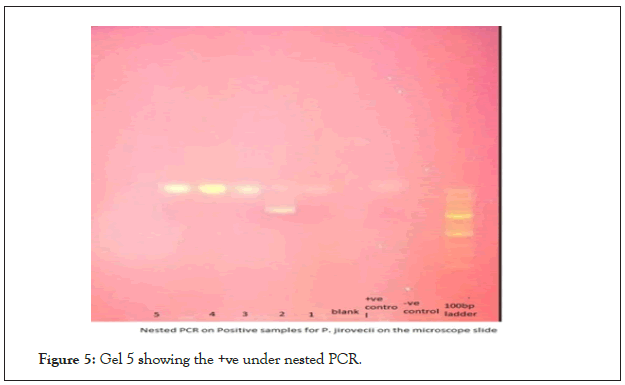
Figure 5: Gel 5 showing the +ve under nested PCR.
The 2nd gel in Figure 5 respectively where the above conditions were followed for the conditions and the gels prestained with ethidium bromide and viewed under the Trans illuminator. In all the gel runs the 100 bp ladder was used followed by the negative control and a positive control. A total of 29 samples out of the total 100 expressed the 364 band fragment on the agarose gel.
The mitochondrial Ribosomal RNA large subunit gene is currently used as a PCR target for PCP diagnosis [22-25]. MtLSU-rRNA, is one of the highly sensitive and specific genetic regions because they are multiple copies of genomes in it [26].
The MtLSU rRNA gene is involved in basic metabolic functions and is considered to be a highly informative locus. This is also the locus used most widely for PCR-based detection of P. jirovecii, as it is present in a large number of copies. To date, seven different MtLSU rRNA sequences have been identified for P. jirovecii, meaning that those sequences are useful for investigating intra specific differences among populations.
Pnuemocystis pneumonia has been shown to be a co-infecting pathogen with other respiratory pathogens with a prevalence of (37%) [27,28]. However the majority of this study specimens were negative for P. jirovecii, These findings highlight the necessity and the importance of fungal diagnosis in high risk population especially those with pulmonary TB. According to this study chi-squire test, there was significance of co-infection Pneumocystis jirovecii with Mycobacterium tuberculosis with p-value of 0.86, different from the study done by Chakaya et al.,[27] and Bii et al.,[28].
This study demonstrated that expectorated sputum can be used for the reliable detection of P. jirovecii, especially when used in conjunction with sensitive PCR technology. Toluidine blue stain detected P. jirovecii cysts in 27.17% of the patients. The Nested PCR detected 38% positive for P. jirovecii. The sensitivity and specificity of nested PCR as compared to TBO staining was 4.27% and 20.6% respectively. Nested PCR test detected additional 12 more patients than toluidine O staining technique showing high sensitivity, TBO had an acceptable sensitivity and very high specificity. P. jirovecii was detected in 29 patients by nested PCR and in 17 patients by TBO microscopy, it was demonstrated that microscopy was about 25% less sensitive compared with a combination of methods that did not include PCR. If this approach had been applied to our patients with positive PCR results, TBO should have diagnosed 29 cases rather than 17 [29,30].
A positive TBO stain generally indicated PCP, so this may be considered the minimum frequency of infection (3.6%). Simultaneous co-infection with M. tuberculosis and P. jirovecii is not thought to be common, which may explain why the prevalence of P. jirovecii appeared to be low in regions with high M. tuberculosis burdens [31-35].
PCP was detected in three of the patients with other respiratory pathogens, two of whom were infected with C. albicans and one with yeast other than C. albicans. The presence of PCP may indicate colonization of these patients; however, it was more likely that Candida was a co-existing oral pathogen in patients with PCP, or at least in those who were HIV-positive, because oral candidiasis is common in these patients [36-40].
Over the past decade, studies have indicated that PCP may be more frequent in TB smear-negative patients. The blurred picture of PCP prevalence in Africa may be due to empirical therapy and prophylaxis using cotrimoxazole in HIV/AIDS patients [41-46].
The Data obtained from this study is an indication of the co-infection of fungi and pulmonary tuberculosis. The study signifies the need of a mycological evaluation of non-healing pulmonary tuberculosis and prudent antifungal treatment based on the culture results rather than depending on broad spectrum antibiotics for cure. Induced sputum is more cost-effective; however, it is also of limited use because it requires staff training, patient preparation and dedicated rooms. A few studies have compared the value of induced sputum with expectorated sputum and they concluded that the difference in yield was negligible. The current study demonstrated that expectorated sputum can be used in resource-limited settings in sub-Saharan Africa for the routine diagnosis of PCP.
Pneumocystis jirovecii is fastidious and thus in vitro culture is difficult. As Pneumocystis remains a non-cultivable microorganism, traditional diagnostic methods rely on microscopic observations of the pathogen in respiratory specimen which is inherently non-specific.
The research was supported by project on “Consortium platform on Vaccines and diagnostics”, Indian Council of Agricultural Research, Government of India, New Delhi, India. I acknowledge my supervisors for their critical analysis of my work and their constant encouragement. Secondly, I acknowledge the Centre for Microbiology Research (CMR), KEMRI, for the provision of laboratory space where most of my work was done. I also acknowledge the Coast General Hospital, Mombasa for their support in acquiring samples from their hospital. I also want to acknowledge the laboratory staff of CMR (KEMRI) for their technical support. Finally I acknowledge my colleagues, friends and family for their constant support.
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
[Crossref] [Google scholar] [PubMed]
Citation: Saitoti A, Wamunyokoli F, Njerwanna SL, Christine Bii (2022) Pneumocystis jirovecii in TB: Smear-Negative and Retreatment Cases at the Coast General Hospital. Virol Mycol. 11:246.
Received: 25-Oct-2022, Manuscript No. VMID-22-19818; Editor assigned: 27-Oct-2022, Pre QC No. VMID-22-19818 (PQ); Reviewed: 10-Nov-2022, QC No. VMID-22-19818; Revised: 17-Nov-2022, Manuscript No. VMID-22-19818 (R); Published: 25-Nov-2022 , DOI: 10.35248/2161-0517.22.11.246
Copyright: © 2022 Saitoti A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.