Research Article - (2022)Volume 8, Issue 5
Prevalence of Multidrug-Resistant Bacterial Isolates from Chickens, Goats, Cattle and Pigs in Bvumbwe, Malawi
Martin H Kalumbi1*, Zefaniah J Katuah1, Atusaye E Nyirenda1, Blessings Katiniche1, Robert Chinyama1, Donita Moyo1, Simon Thugo1, Madalitso Mlozen2, Chikondi Kamwendo2, Elias Bonya2, Adam M Nyanda2, Jonathan Majamanda3, Wilfred Taika3, Patrick Chagwa3 and Linly Linje3Abstract
Background: Antibiotic resistance is a serious problem worldwide affecting human and animal health. There is a potential that antibiotic resistant bacterial strains are passed on from animals to humans leading to more serious bacterial infections and burden.
Aim: The aim of this study was to determine antibiotic resistant bacterial profile in chickens, cattle, goats and pigs.
Methods: Prospective cross sectional laboratory based study was conducted on droppings, mouth, nose and hooves samples obtained from chickens, cattle, goats and pigs from Bvumbwe, Malawi. Gram stain and biochemical reactions were used to identify bacterial pathogens. Susceptibility of bacterial isolates to commonly used antibiotics in Malawi was done using disk diffusion method under both aerobic and anaerobic conditions.
Results: In total, 110 animal samples were examined and all (100%) were found positive with at least one type of bacteria. Citrobacter, S. aureus, Bacillus, E. coli, Clostridium, Klebsiella, Streptococcus, other coliforms and Staphylococcus spp. were isolated. Bacillus spp. recorded the highest prevalence (77.3%), followed by Citrobacter spp. (41.6%) and S. aureus (39.1%). S. aureus and Citrobacter spp. demonstrated multidrug resistance to at least four antibiotics including Gentamycin, Tetracycline, Ampicillin, Ciprofloxacin, Chloramphenicol and Erythromycin. Highest resistance of 41.7% was observed in S. aureus followed by Citrobacter species of 33.3%. Among the antibiotics tested, highest resistance was portrayed by Ampicillin (77.8%) and Tetracycline (66.7%).
Conclusion: This study highlighted that healthy farm animals such as chickens, cattle, goats and pigs harbour multidrug-resistant bacteria with high levels of Ampicillin and Tetracycline resistance. This will likely limit options for antibiotic therapy in animals and humans. Efforts are therefore needed to control the use, distribution, storage and sale of antibiotics in veterinaries.
Keywords
Antibiotic resistant bacterial profile; Human and animal health; Susceptibility testing; Bacterial infections
Introduction
Antibiotic resistance is a serious problem worldwide affecting human and animal health. Some bacteria are resistant to more than one antibiotic class and this multidrug-resistance is one of the top 10 threats to global health [1]. Development of multidrug- resistant bacteria could be due to improper use of antibiotics in both humans and animals [2]. Antibiotic resistance has been reported in Malawi and across Sub-Saharan Africa [3,4]. In this region, there is a potential that antibiotic-resistant bacterial strains are passed on from animals to humans and vice versa through food chain and direct contact. This spread of antibiotic-resistant strains could lead to narrowed therapeutic options, more serious bacterial infections and burden and exerting pressure to already struggling health systems in these developing countries. It is estimated that by 2050, antibiotic-resistant infections are likely to cause the death of 10 million people per annum if the current trends of antimicrobial resistance persist [5,6].
Currently, there is a growing demand for high quality animal protein in Low and Middle Income Countries (LMICs) such as Malawi due to high levels of malnutrition and communicable diseases. This demand has been associated with an increase in livestock farming. In most cases, livestock is free-roaming and gets into contact with contaminated water, food and infected human beings. To protect or treat such animals, farmers have resorted to the use of antibiotics intended for human use and this has been linked to the development of multidrug-resistant pathogens in animals and humans [7,8]. The rise of antibiotic resistance in zoonotic pathogens poses a serious challenge to humans and animals as it is associated with fatal untreatable infections [9].
The accumulation of harmful resistant bacteria in livestock and poultry is primarily attributed to the frequent use of inadequate doses of antibiotics in livestock farming such as chickens, goats, pigs and cattle [9]. It is reported that the use of antibiotics in food producing animals outweigh human consumption in many countries [10]. In animals, antibiotics are used as growth promoters and are inappropriately used as low-cost substitutes for hygienic measures that are aimed at infection prevention.
This indiscriminate use of antibiotics in food producing animal farming facilitates development of antibiotic resistance in pathogenic and commensal bacteria. These bacteria use different mechanisms in development of resistance such as enzymatic inhibition, Penicillin-Binding Protein (PBP) modifications, porin mutations, efflux pumps, and target changes [11]. Apart from the reported concerns linked to emergence of antibiotic resistance in bacteria isolated from livestock and poultry, there are also human health concerns associated with the presence of antibiotic residues in meat and eggs [12]. This could be one of the contributing factors to high levels of Bacterial Blood Stream Infections (BSIs) and mortality in Sub-Saharan Africa [13]. These bacterial blood stream infections are usually difficult to treat as the pathogens are exposed to antibiotic residues or compounds prior to prescribe treatment. Enterobacteriaceae are some of the major pathogens that have developed resistance to third generation Cephalosporins leading to widespread reliance on Ceftriaxone for management of sepsis in Africa, including Malawi. In addition to Cephalosporins, others are resistant to Methicillin, Tetracycline, Enrofloxacin, Streptomycin, Neomycin, Gentamicin and Amoxicillin among others [14,15].
In Malawi, chickens, cattle, goats and pigs are predominately used as a source of income and food although they are key reservoirs of bacteria. Malawi and other governments established regulations through different institutions such as Malawi Bureau of Standards (MBS) to protect consumers from harmful effects of pathogenic bacteria [16,17]. Unlike with other countries, the enforcement of these regulations in LMICs including Malawi is challenged by numerous factors such as poor examination, inspection, analysis and testing before human consumption. Currently, there are no published antibiotic resistance profiles and cross contamination studies that have been reported on chickens, cattle, goats and pigs in Malawi.
In this regard, the current study was therefore carried out to assess the presence of bacteria in the aforementioned farm animals and determine resistance patterns of isolated bacteria to commonly used antibiotics in Malawi’s hospitals.
Materials and Methods
Study design
The study employed laboratory based experimental design. This involved isolation, identification and antibiotic susceptibility testing of bacteria from chickens and animals belonging to bovidea family namely; goats, cattle and pigs against commonly used antibiotics in Malawi.
Study site
The study was conducted in Bvumbwe, an agriculture-based rural settlement located approximately 21 km from east of Blantyre city and laboratory analyses were performed at Malamulo College of Health Sciences in Thyolo District located in the Southern Region of Malawi. Bvumbwe was chosen based on its rural setting and highly populated with large number of food producing animal farmers. Anecdotal evidence shows that majority of families are either subsistence or commercial animal farmers and are the major producers of meat and milk in the district and surrounding districts.
Sampling of study animals
In March 2021, the samples were obtained from four different farmers who were selected using simple random sampling. Sampling of herds and flock of chickens were based on the willingness of farmers and number of animals being kept. For instance, only farmers with herd and flocks not less than 15 were selected. In total, 110 samples were collected from mouth, nose and hooves of cattle, pigs and goats without any sign and symptom of bacterial infection while droppings and rectal swabs were collected from healthy chickens. Animals from kraals containing both healthy and diseased animals were excluded from this study. The samples were not collected repeatedly from these farm animals.
Sample collection and transportation
ESwab™ (Medline industries, China) sterile swabs were used to swab the mouth, nose, hooves, rectum and droppings of study animals using sterile disposable gloves. The swabs containing the samples were carefully labelled and placed in a 2 ml nutrient broth agar transport media which were packed in Ziploc bags. Then after, the swabs were packed in a cooler box at a temperature range of 4 to 8°C and transported to Malamulo College of Health Science Microbiology Laboratory for analyses. The samples were immediately prepared and analysed at the laboratory.
Bacterial culture, isolation, and identification
All the laboratory media preparations were done according to standard procedures [18]. The swabs were removed from the transport media and streaked over the petri dishes (plates) containing well prepared nutrient, blood, MacConkey and Salmonella or Shigella agar. The plates were labelled and incubated aerobically and anaerobically at 37°C for 24 hours. Colony characteristics were noted and the smears were prepared from different colonies on glass slides. The smears were labelled and subjected to Gram’s reaction for cell morphology identification using 100x light microscope. Conventional biochemical tests were performed to identify the bacterial isolates further [19]. Biochemical tests which were done include catalase, coagulase, indole, oxidase, simon citrate, urease and triple sugar iron tests (Scharlau, Germany).
Antibiotic susceptibility tests
Disk diffusion (modified Kirby-Baeur) method was employed to determine antibiotic resistance and was done according to Clinical and Laboratory Standards Institute [20]. The tests were done on Mueller-Hinton agar (Oxoid, Hampshire, England). About 12 commercially available antibiotic susceptibility disks (Oxoid, England) were used. The disks were Amikacin (30 μg), Amoxicillin (25 μg), Bacitracin (10 μg), Ceftriaxone (30 μg), Penicillin (10 μg), Doxycycline (30 μg), Ampicillin (10 μg), Tetracycline (30 μg), Erythromycin (15 μg), Gentamycin (10 μg), Chloramphenicol (30 μg) and Ciprofloxacin (5 μg). Aforementioned antibiotics were selected following consultations with different local hospitals and they are currently used in hospitals in Malawi. The pure isolates and the controls (reference strains) were inoculated on a sterile Muller-Hinton agar and incubated aerobically and anaerobically for 24 hours at 37°C. The reference strains were obtained from Malamulo Mission Hospital and were characterised by the hospital with known susceptibility profiles to various antibiotics. The zones of inhibition were measured using a ruler in millimetres and interpreted as either susceptible or resistant following Clinical and Laboratory Standards Institute guidelines [20-22].
Statistical analysis
Data was analysed using Statistical Package for Social Science (SPSS, version 20, Chicago: IBM Corp.) and Microsoft Excel (version 16, Microsoft Corporation, New York, USA). Laboratory analyses were performed in triplicates and the means were computed. The data were summarised and presented in tables and figures. The prevalence of bacterial isolates in different samples was computed by dividing positive samples by total number of examined samples and multiplied by one hundred.
Ethical approval
Permission to conduct the study was sought from National Health Science Research Committee (NHSRC), Department of Biomedical Sciences at Malamulo College of Health Science and Bvumbwe Veterinary Officer. An informed oral consent was obtained from the owners of the study animals during sample collection. All the methods were performed in accordance with International Council for Laboratory Animal Science (ICLAS) guidelines.
Results
Prevalence of bacterial isolates
A total of 110 samples were collected from chickens (n=10), cattle (n=30), goats (n=40) and pigs (n=30). Bacteria were detected in all the 110 (100%) samples (Table 1 and Figure 1). Nine types of bacteria isolated from these food producing farm animals as shown in Table 1. Among the samples, the most predominant bacterial isolates were Bacillus spp. (77.3%), followed by Citrobacter spp. (41.6%) and Staphylococcus aureus (39.1%). On the other hand, the least predominant bacterial isolates were Streptococcus spp. (1%). Of all the farm animals, chickens harboured six types of bacteria while the rest of the animals harboured at most five types.
| Isolated bacteria | Prevalence in different samples [n (%)] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chickens (n=10) | Cattle (n=30) | Goats (n=40) | Pigs (n=30) | Total (%) | ||||||
| Mouth | Nose | Hooves | Mouth | Nose | Mouth | Nose | Hooves | |||
| Citrobacter sp | 3(30) | 10(100) | 10(100) | 5(50) | 2(10) | 0 | 6(60) | 6(60) | 4(40) | 46(41.8) |
| Staphylococcus sp | - | 4(40) | 1(10) | 10(100) | 2(10) | - | 0 | 6(60) | 0 | 23(20.9) |
| S. aureus | 3(30) | - | - | - | 20(100) | 20(100) | - | - | - | 43(39.1) |
| Bacillus sp | 3(30) | 10(100) | 8(80) | 8(80) | 20(100) | 20(100) | 2(20) | 10(100) | 4(40) | 85(77.3) |
| E. coli | 2(20) | - | - | - | - | - | 0 | 0 | 6(60) | 8(7.3) |
| Other coliforms | 1(10) | - | - | - | - | - | 8(80) | 0 | 4(40) | 13(11.8) |
| Clostridium sp | - | - | - | - | 1(5) | 16(80) | - | - | - | 17(15.5) |
| Klebsiella sp | - | 0 | 5(50) | 0 | - | - | - | - | - | 5(4.5) |
| Streptococcus sp | 1(10) | 2(20) | 0 | 0 | - | - | - | - | - | 3(2.7) |
Table 1: Prevalence and distribution of bacteria isolated from chickens, cattle, goats and pigs.
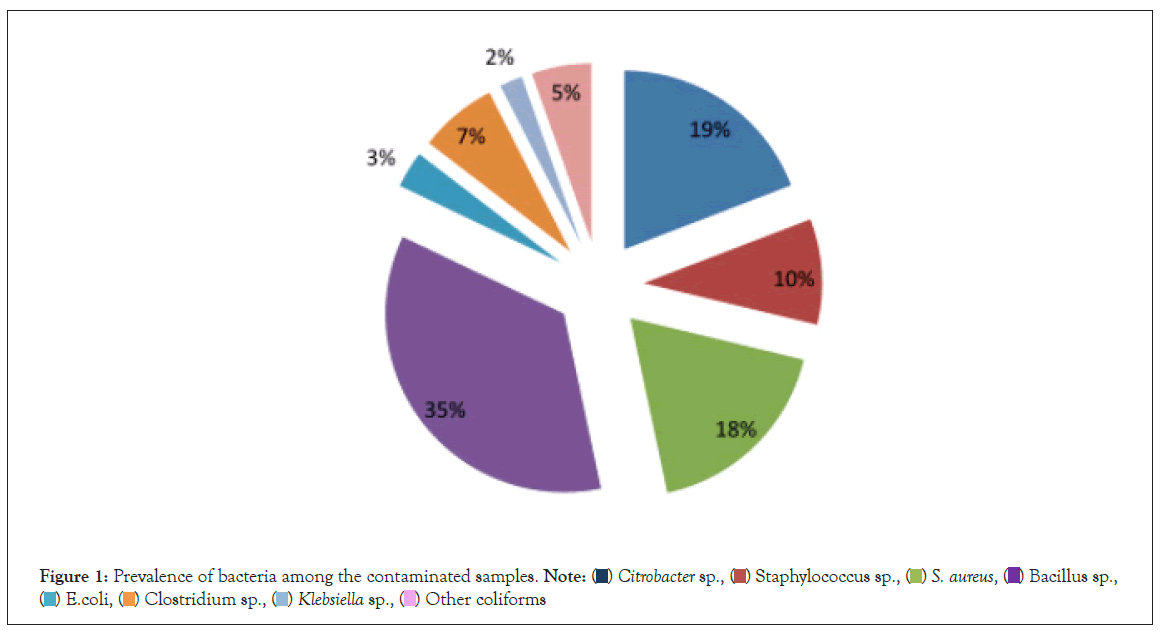
Figure 1: Prevalence of bacteria among the contaminated samples. 
Antibiotic susceptibility
Susceptibility profiles of bacterial isolates to antibiotics commonly used to treat human infections in Malawi are summarised in Table 2. The levels of antibiotic resistance ranged from 0 in Streptococcus spp. to 41.7% in S. aureus. The highest levels of resistance were observed in S. aureus (41.7%) followed by Citrobacter spp. (33.3%) while Bacillus spp and E. coli demonstrated similar moderate levels of antibiotic resistance of 25%. These bacterial isolates were resistant to at least three classes of antibiotics. Among the antibiotics, the isolates were highly resistant to Ampicillin (77.8%) followed by Tetracycline (66.7%). The resistance pattern to Ampicillin and Tetracycline was more common in cattle and pigs.
| Bacterial isolates | Antibiotic resistance profile | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chickens | Goats | Cattle | Pigs | n (%) | |||||
| Resistance | Intermediate | Resistance | Intermediate | Resistance | Intermediate | Resistance | Intermediate | ||
| Citrobacter sp | - | CIP | PEN | TET | AMP-CHLO-TET | ERY | CHLO-TET | AMP-ERY | 4 (33.3) |
| Staphylococcus sp | - | - | - | - | - | - | - | - | 0 |
| S. aureus | - | GEN-TET-AMP | STRE-PEN | - | AMP-GEN | TET-CIP | AMP-TET | GEN | 5 (41.7) |
| Bacillus sp | - | GEN-ERY | ERY-TET | - | AMP-TET | CIP-ERY | AMP-TET | ERY | 3 (25) |
| E. coli | GEN | ERY-TET | - | - | - | - | AMP-TET | - | 3 (25) |
| Other coliforms | - | - | - | - | - | - | AMP-TET | - | 2 (16.7) |
| Clostridium sp | - | - | AMP | TET | - | - | - | - | 1 (8.3) |
| Klebsiella sp | - | - | - | - | AMP-TET | - | - | - | 2 (16.7) |
| Streptococcus sp | - | AMP | - | - | - | AMP-CHLO-GEN-ERY | - | - | 0 |
Note: CIP: Ciprofloxacin; PEN: Penicillin; TET: Tetracycline; AMP: Ampicillin; CHLO: Chloramphenicol; ERY: Erythromycin; GEN: Gentamycin; STRE: Streptomycin; PEN: Penicillin; -: blank; n: number of positive resistance (each drug counted once within a row).
Table 2: Susceptibility profiles of bacteria isolated from healthy chickens, goats, cattle and pigs.
Discussion
This study applied microbiological and epidemiological approaches to characterise the epidemiology and antimicrobial susceptibility of different species of bacteria isolated from chickens, cattle, goats and pigs. The prevalence of bacteria reported herein agrees with the theory that postulates that food producing farm animals are constantly infected by different bacteria [14]. In this study, Citrobacter, Staphylococcus, Bacillus , Escherichia, Clostridium, Klebsiella and Streptococcus species, and other coliforms were isolated from chicken, cattle, goat and pig samples. Although aforementioned organisms were isolated, their prevalence and distribution among the samples were inconsistent, both high and low prevalence were recorded in Bacillus spp and Streptococcus spp respectively. This variation may have resulted from several factors such as (1) collection of samples from different kraals and animals. Different kraals are subjected to different hygienic conditions and those poorly managed kraals are likely to harbour a lot of microorganisms. (2) Genetic variation of animals. The widely accepted idea is that there is genetic variation in different animals, making the host resistant to bacterial attack and survival. (3) Differences in feeds and their preparations. The fact that these samples were collected from various animals and different kraals their feeds are more likely to be different as noted during sample collected. These feeds may be contaminated by different bacteria from different locations in the community and would favour growth of distinctive bacteria. (4) Use of antibiotics. Some populations of bacteria may have been eliminated by antibiotics used by the owner of the farms. And (5) survival mechanisms of bacteria in host bodies; whereas it is possible that some bacteria would fail to survive in bovine animals. For instance, the presence of Bacillus species in all the samples is attributed to its ability to produce spores in times of stress conditions and vegetate when the conditions are favourable. As such, the present study suggests that distribution of bacteria in different samples and animals from the same community is heterogeneous. Although the study did not endeavour to systematically identify the actual species of some bacteria using Polymerase Chain Reaction (PCR), it is our recommendation that future studies take that path. Several studies have documented similar isolated bacteria in different farm animals globally, but none has matrixed a combination of chickens, cattle, goats and pigs [14,21,23]. Variation in the types and number of bacteria isolated in different studies is attributed to differences in location, feed and type of samples tested.
Over six types of bacteria were isolated from different farm animals. The occurrence of types of bacteria was high in Chickens where 66.7% of these types of bacteria were isolated. Types of bacteria reported herein are an indication of the risk of exposure of these chickens and other farm animals to bacteria. In fact, the results indicates that the reasons could be multifactorial as it became evident during visual assessment of the kraals and feeds that hygiene was not satisfactory. For instance, there was high animal density and not well cleaned surfaces. Similar factors were reported in which bacteria contamination was associated with animal density and scrapings, manure storage, water and pest control among others [24].
An unusual antibiotic resistant bacterial profile in chickens, cattle, goats and pigs were recorded in this study. High multidrug resistant profiles to commonly used antibiotics tested were demonstrated by S. aureus and Citrobacter spp. in cattle, goats and pigs. For instance, Ampicillin resistance was common in these bacteria followed by Tetracycline, Gentamycin and Chloramphenicol. This emergency of multidrug resistance could be due to the improper use of antibiotics in the farms as indicated orally by the farmers that they use antibiotics to treat their animals. Others had mentioned that they do so without consulting veterinary officers. This implies that farmers are not following WHO guidelines on the use of antibiotics in animal production [25]. It could also be due to the fact that the owners employ free range system of animal keeping especially after harvesting of crops and also animals were exposed to other chemicals with compounds similar to antibiotics that could confer cross resistance.
Occurrence of similar resistance profile in pigs for S. aureus, Bacillus spp, E. coli and other coliforms against Ampicillin and Tetracycline re-affirms that the owners of the pigs were improperly administering these antibiotics to the pigs. Similarly, in this area the owner of the cattle had been using Ampicillin and Tetracycline and the observed resistance was not surprising. It is not surprising that Tetracycline resistance was displayed by these organisms as similar results were observed previously in Malawi’s neighbouring countries namely, Tanzania and Zambia [25]. In these countries, organisms isolated from cattle, poultry and pig samples were resistant to Tetracycline. This similarity could be due to free flow of animals among these countries in trade and pasturing hence easy transfer of resistance genes and organisms. It is now clear that in these three countries Tetracycline has been abused in animal farms.
The resistant strains are not only common in Africa as mentioned earlier but also in other continents where farming has been exploding. In Malaysia, similar patterns of resistance for S. aureus but not for E. coli have been reported in samples collected from diseased farm animals [14]. However, the findings from the current study have been novel as the study was done among physically healthy animals. Just like in humans, the data from this study clearly indicate that farm animals may look physically healthy but carrying bacteria with resistance genes. This has a strong impact in both food security and safety as these resistant organisms can be passed on to humans through food consumption or contact with infected environment. As such, this pattern of resistance to commonly used antibiotics in local hospitals is a great threat to Low and Middle Income Countries with limited variety of antibiotics. This serves as a wakeup call to all farmers and Ministry of Agriculture to step up measures that should protect consumers such as strong surveillance of antimicrobial resistance in agriculture and proper use of antibiotics. Further, it is strongly recommended that distribution, handling, storage and sale of antibiotics in Malawi and other Sub- Saharan countries should be effectively monitored and controlled. These measures can help in limiting emergence and distribution of resistant strains yet protecting the citizens who are not aware of health risks associated with such bacterial strains.
Conclusion
This study provides the first published data on the profile of antibiotic resistant strains isolated from chickens, cattle, goats and pigs in Malawi. The study has revealed that the most common bacterial isolates are Citrobacter, S. aureus, Bacillus, E. coli, Clostridium, Klebsiella, Streptococcus, other coliforms and Staphylococcus species. S. aureus and citrobacter species demonstrated highest multidrug resistance while Ampicillin and Tetracycline resistance was the most distributed resistance among the samples. The data presented herein suggests that mitigation efforts are needed to limit emergency and distribution of resistant bacterial strains in the farm animals as a way of addressing the challenge of antibiotic resistance in Malawi and globally.
References
- World Health Organization. Ten threats to global health in 2019
- Pokharel S, Shrestha P, Adhikari B. Antimicrobial use in food animals and human health: Time to implement one health approach. Antimicrob. Resist. Infect. Control. 2020; 9(1):1-5.
- Haigh K, Dube Q, Kasambara W, Feasey NA, Lester R. Cephalosporin resistance in Malawi. Lancet Infect Dis. 2020;20(3):285-286.
- Essack SY, Desta AT, Abotsi RE, Agoba EE. Antimicrobial resistance in the WHO African regioncurrent
status and roadmap for action. J Public Health. 2017;39(1):8-13. - O'Neill J. Tackling drug-resistant infections globally: final report and recommendations.
- Oâ??Neill J. Review on antimicrobial resistance. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. 2014.
- Aarestrup FM, Kruse H, Tast E, Hammerum AM, Jensen LB. Associations between the use of antimicrobial agents for growth promotion and the occurrence of resistance among Enterococcus faecium from broilers and pigs in Denmark, Finland, and Norway. Microb Drug Resist. 2000;6(1):63-70.
- O'Neill J. Antimicrobials in agriculture and the environment: reducing unnecessary use and waste. 2015
- Aarestrup FM. The livestock reservoir for antimicrobial resistance: a personal view on changing patterns of risks, effects of interventions and the way forward. Philos Trans R Soc Lond B Biol Sci. 2015.
- Center for Veterinary Medicine. CVM Updates-FDA Annual Summary Report on Antimicrobials Sold or Distributed in 2013 for Use in Food-Producing Animals.
- Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2):4-2.
- Agyare C, Boamah VE, Zumbi CN, Osei FB. Antibiotic use in poultry production and its effects on bacterial resistance. 2018:33-51.
- Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49.
- Haulisah NA, Hassan L, Bejo SK, Jajere SM, Ahmad NI. High levels of antibiotic resistance in isolates from diseased livestock. Front Vet Sci. 2021.
- Kimera ZI, Mshana SE, Rweyemamu MM, Mboera LE, Matee MI. Antimicrobial use and resistance in food-producing animals and the environment: an African perspective. Antimicrob Resist Infect Control. 2020 Dec;9(1):1-2.
- Malawi Bureau of Standards Catalogue of Malawi standards. Blantyre, Malawi: Malawi Bureau of Standards.2017.
- Malawi Government The Malawi Government Gazettee â?? Malawi Bureau of Standards Act 2012. Zomba, Malawi: Malawi Government.2014.
- Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, Fitzpatrick E. Veterinary microbiology and microbial disease. JWS;2011.
- Collee JG, Miles RS, Watt B. Tests for identification of bacteria. Mackie and McCartney practical medical microbiology. 1996;14:131-49.
- Wayne PA. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing.2020.
- Arbab S, Ullah H, Wang W, Li K, Akbar A, Zhang J. Isolation and identification of infection-causing bacteria in dairy animals and determination of their antibiogram. J Food Qual.2021.
- Chigerwe M, Mavangira V, Byrne BA, Angelos JA. Antibiotic resistance patterns of bacteria isolated from indwelling Foley catheters following tube cystostomy in goats with obstructive urolithiasis. J Vet Diagn Investig. 2017;29(3):316-20.
- Langata LM, Maingi JM, Musonye HA, Kiiru J, Nyamache AK. Antimicrobial resistance genes in Salmonella and Escherichia coli isolates from chicken droppings in Nairobi, Kenya. BMC Res Notes. 2019;12(1):1-6.
- Ghougal K, Moreno Roldán E, Espigares RodrÃguez E. Risk factors related to bacterial contamination by Enterobacteriaceae and fecal coliforms and the prevalence of Salmonella spp. in Algerian farms, slaughterhouses and butcheries: a two-year follow-up study. AIMS.
- Aidara-Kane A, Angulo FJ, Conly JM, Minato Y, Silbergeld EK, McEwen SA, et al. World Health Organization (WHO) guidelines on use of medically important antimicrobials in food-producing animals. Antimicrob Resist Infect Control. 2018;7(1):1-8.
Author Info
Martin H Kalumbi1*, Zefaniah J Katuah1, Atusaye E Nyirenda1, Blessings Katiniche1, Robert Chinyama1, Donita Moyo1, Simon Thugo1, Madalitso Mlozen2, Chikondi Kamwendo2, Elias Bonya2, Adam M Nyanda2, Jonathan Majamanda3, Wilfred Taika3, Patrick Chagwa3 and Linly Linje32Department of Biomedical Science, Biochemistry Research Group, Malamulo College of Health Science, Malawi Adventist University, P.O Box 55, Makwasa, Malawi
3Department of Biomedical Science, Parasitology Research Group, Malamulo College of Health Science, Malawi Adventist University, P.O Box 55, Makwasa, Malawi
Citation: Kalumbi MH, Katuah ZJ, Nyirenda AE, Katiniche B, Chinyama R, Moyo D, et al. (2022) Prevalence of Multidrug-Resistant Bacterial Isolates from Chickens, Goats, Cattle and Pigs in Bvumbwe, Malawi. Appli Microbiol Open Access. 8: 234.
Received: 08-Jul-2022, Manuscript No. AMOA-22-18272; Editor assigned: 11-Jul-2022, Pre QC No. AMOA-22-18272(PQ); Reviewed: 27-Jul-2022, QC No. AMOA-22-18272; Revised: 04-Aug-2022, Manuscript No. AMOA-22-18272(R); Published: 11-Aug-2022 , DOI: 10.35284/2471-9315.22.8.234
Copyright: © 2022 Kalumbi MH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.