Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2019)Volume 12, Issue 4
Background: Plant fungal diseases are the primary causes of foliage and crop loss eventually affecting the overall economic outcome and yield quality. Hence, various chemical compounds are employed to eradicate the fungi in agriculture.
Methods: Virtual screening and molecular docking strategies provide themselves as great alternatives to find lead compounds. Lead compounds for each fungal infection was docked to target protein sequence and assessed for the strongest interaction.
Findings: Various molecules were taken under the study, for being the target ligands to bring about a fungicidal reaction in the plant pathogen system. The screening of molecules was done thoroughly to produce the results. Ligands identified through this study allow us to make plant host fight against the fungal pathogen and prevent the occurrence of the disease. The interactions have been thoroughly studied with various softwares like SPDBV and PyMol and through various online databases like STRING, GenePept, PDB, UniProt, PatchDock, Protein structure prediction server -2 and others for the overall evaluation of the drug molecule designed and to study its overall effects for the overall higher efficacy and to prevent the occurrence of the fungal disease and management of the fungal pathogens in agriculture against various economically valuable plants. The lead compounds revealed several hydrophobic and polar contacts were demonstrated by comparing interactions.
Applications: The molecular affinity of the fungicidal compound has been tested against the target pathogen as well as the host system components to understand the interaction and to draw out the functioning and the analysis. The compatibility between the molecule and the protein has been studied to decipher the effectiveness of the molecule and its effects in the system. The present results let us establish lead compounds that can be used for the development of antifungal drugs although structural activity relationship studies have to be undertaken.
Fungi; Fungicide; Agriculture; Molecular docking; Molecules
Managing fungal infections or diseases that economically impact plant yield and quality can be managed by the use of fungicide which specifically inhibits or kills or stall the growth of the fungus causing the disease [1,2]. Fungicides are also used to control the disease during various stages including establishment and development of a crop, increase in productivity, reduction in the residual infection, and improve the storage life and the quality of harvested plants [3]. According to the target sites, commercially available agricultural fungicides are classified by the international Fungicide Resistance Action Committee (FRAC). However, this classification does not include metalloorganic, inorganic and human hazardous fungicides. The emergence of resistant fungal strains, difficulty in the treatment and the multi-fold increase in the number of fungal infections necessitates and prioritises the discovery of new molecular scaffolds to achieve effective control. The urgency in dealing with fungal infections is reflected by pharmaceutical companies creating a division for pesticides especially for the agrarian market as agricultural fungicides are an excellent source of lead structures. Computation-aided drug designing can help design lead molecules for target biological functions and decipher a functional overlap in molecular target sites or target similar processes or molecules [4]. Structurally, a fungicide has a specific target site where it acts to disrupt a biochemical process or function. If there is an alteration in the target site, the fungicide can no longer bind or can bind with low affinity and is unable to exert its toxic effect. Molecular docking is an in-silico technique that can be used to model the interactions between the fungicide molecules and the target protein and help understand the role of changes if associated, explain the variations in the toxicity of the molecules with or without the same mode of action and help design new inhibitors with greater affinity to the binding site [5-7]. In this study, major fungal pathogens that contribute to a large percentage of plant disease with a wide host range of economic plants were identified. Target molecule identified as ligands with their target proteins were confirmed by a literature search. The functioning and configuration of the molecule was identified using the energy profile and simulated with all possible conformations and orientations. The overall design of the study was designed to predict the molecules effective against the fungi and not against the host plant, thereby negating the bioavailability of the fungicide for the plant.
Identification of targets
The major fungal pathogens that contributed to a large percentage plant disease were identified and selected with their target host plants that included the fungi belonging to various classes and genera: Alternaria, Puccinia, Botrytina, Uromyces, Phytophthora, Melampsora and Magnaporthe, that infect a wide range of economic plants reducing their overall yield [8]. Target molecules were identified that could induce resistance/ prevention from these plant pathogens. Some of these molecules were pre-existing chemical fungicides while others functioned as the elicitor molecules to induce the resistance response in host system [9]. The list of identified target molecules of corresponding fungi has been listed in Table 1.
| Pathogen | Disease caused | Fungicide | Ligand | Protein |
|---|---|---|---|---|
| Fusarium | Fusarium wilt | Chitosan | Chitosan | cch1 |
| Magnaporthe | Rice blast | Probenazole | Probenazole | peroxidase |
| Probenazole | polyphenoloxidase | |||
| Phytophthora | Late blight of potato | Mancozeb | Mancozeb | cytochrome-c-oxidase |
| Botrytina | Necrotrophic | Pyrimethanil | Pyrimethanil | cystathionine-β-lyase |
| Alternaria | Early blight of Solanaceae members | Mancozeb | Mancozeb | monooxygenase |
| Melampsora | Flax rust | Imidazole | Imidazole | demethylase |
Table 1: Major pathogens, their respective fungicide, target proteins and identified ligand molecules of the present study.
Structural and functional analysis of ligands and target protein
The structures of these reported molecules were identified and analyzed using the PDB and PubChem- NCBI structure viewer and constructed for further work using Chemsketch [10]. The structural availability allows understanding of the interaction as well as of the chemical; nature of the compound. Through the literature databases like PubMed and PMC the proteins in the host system which interact with target ligands were identified, as mentioned in Table 2. The structures of these proteins were further elucidated after using NCBI-GenPept and Protein structure prediction server respectively. The energy profiles of these proteins were screened using the Swiss PDB viewer SPDBV; [11] and the minimized energies of the protein molecules were elucidated. The different forms of energy that encompass the total energy of the molecule were analysed separately in the regular and the minimized state to understand the functioning and the configuration of the molecule. Further, the structural components of the individual protein structures were elucidated using the SOPMA software (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html) that allowed us to decipher various structural components and the overall protein profile of the reported proteins known to be interacting with our target ligand molecules.
| Chain type | Peroxidase | Polyphenol oxidase | Demethylase | CCH1 | Monooxygenase | Cytochrome c oxidase | Cystathionine beta-lyase | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | Number | % | Number | % | Number | % | Number | % | |
| Alpha helix | 136 | 37.88 | 125 | 21.66 | 696 | 41.7 | 939 | 44.63 | 226 | 40.5 | 64 | 27.35 | 189 | 41.18 |
| 310 helix | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pi helix | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Beta bridge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Extended strand | 57 | 15.88 | 109 | 18.89 | 182 | 10.9 | 198 | 9.41 | 99 | 17.74 | 55 | 23.50% | 76 | 16.56 |
| Beta turn | 19 | 5.29 | 18 | 3.12 | 86 | 5.15 | 57 | 2.71 | 36 | 6.45 | 14 | 5.98 | 35 | 7.63 |
| Bend region | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Random coil | 147 | 40.95 | 325 | 56.33 | 705 | 42.24 | 910 | 43.25 | 197 | 35.3 | 101 | 43.16 | 159 | 34.64 |
| Ambiguous states | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00% | 0 | 0 |
| Other states | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00% | 0 | 0 |
Table 2: SOPMA values for various proteins known to be interactive with the identified ligand molecules.
Protein interaction
A single chemical molecule does not interact with just one other molecule in a biological system. Various proteins may possess a structural similarity with the one screened and hence, it is important to understand the networking of the protein in view and its various interactions. This is done using the online STRING database which provides the estimated interactions and similarity based on various parameters. Using the STRING database, different proteins with structural similarity with the reported proteins were short listed and their structure was detected.
Docking analysis
In the present study, the docking is mediated between the target drug ligand molecule with the interacting protein molecule using Patchdock analysis [12] tool that provides online docking interface of the files to be submitted in pdb format as a query. The formatting of the file format was done using Open Babel software that can easily convert the given file format to pdb format for the further proceedings. The results are then provided with multiple combinations, as possible for the particular ligand and protein interaction.
Six fungal species involved with Fusarium wilt, rice blast, late blight of potato, necrotrophy, early blight among members of family Solanaceae and flax rust were selected. The respective fungicide for each of these fungal species were identified based on which the proteins were chosen (Table 1). A single protein was selected for ligands chitosan, pyrimethanil and imidazole were chosen whereas probenazole was targeted with two proteins peroxidase and polyphenoloxidase. Although the same molecule (mancozeb) was identified for both the blight diseases, two proteins were identified as targets: cytochrome-coxidase for late blight of potato and monooxygenase for early blight among Solanaceae members. Docking primarily functions on the shape complementarity and simulation between the molecules with all possible conformations and orientations between the protein and the ligand [13]. Docking refers to interfacial analysis of the two components in a system. It is a molecular modelling technique that allows one to find the most favourable orientation of two interactive molecules favouring the study of molecular interaction between the two entities in a reaction. The affinity of a small molecule in drug designing is often related to the free energy calculations involved in binding. The variations found in this relationship is often equated to the interpretation and activity of organic molecules toward the target of interest [14]. The free energy involved in the binding (ΔG) as observed in most of the compounds vary with respect to target with good binding affinity. The computed values have reflected an overall trend relative to the configuration of the complex and stability estimation (Table 3). Seven selected protein molecules were energy-minimized, a process commonly used in established methods to reduce the overall potential energy of proteins for protein-ligand interaction. Since biological systems are very dynamic and have low potential energies (negative ΔG) for spontaneous interaction, energy minimization help attain a conformation with lower ΔG values so as to be considered close to biological system. Although some select proteins such as polyphenoloxidase, cyatathione-β-lyase, monooxygenase showed a negative ΔG values, they were also subjected to energy minimization similar to the other proteins. The results obtained from docking are used to determine the molecular interactions at atomic level between the ligand and the protein using the software-PyMol. Ramachandaran plots (R-plots) were generated for each complex using the SPDBV and analysed for the amino acid distribution of the complex in the allowed and the disallowed regions (Figure 1). This can be understood through the Table 4 and Figure 2 which provides the information collected from the R-plot. This value allows analysis of the overall stability of the complex in whole. The Ramachandran plots of the models were depicted and compared after refinement. The Ramachandran plot (Figure 2 and Table 4) indicates that some amino acids in the best predicted structure are located at outlier region. Each complex was analysed for the total amino acids in the disallowed region, the amino acids involved including and excluding glycine. The highest number of amino acids in the disallowed region was found for the complex cch1+Chitosan with a total of 126 out of which 106 were amino acids other than glycine. The least number of amino acids were found in the complex polyphenoloxidase + probenazole with four glycine residues involved in the interaction. The stability of the complexes is often reported in the Ramachandran plots with the number of glycine residues as it lacks a side chain and can adopt phi psi angles in all four quadrants of the R-plot. A maximum of 24 glycine residues were present in complex demethylase+imidazole and a minimum of four residues in polyphenolxidase+probenazole complex. The energy minimized values have been tabulated for the reported proteins+ligand complex using SPDBV (Table 5). Further the best docking poses during interaction derived from PyMol has been illustrated in Figure 3. The compounds under the observation have a high binding affinity with the receptors. All the ligands are found to form a strong hydrogen bonding with key residues and no ligand was found to stabilize inside the pocket with or without tremendous interactions with key residues of the protein. A prominent role has been played by the amine group in complexes cystathionine–β-lyase+pyrimethanil and cch1+chitosan. Among the other complexes, hydrogen bonding with key residues inside the pocket is observed to be a key determinant for binding of ligand with active residues. It can be assumed that the rigidity of the structures can also pose as a major factor that leads the ligands to attain docking poses and orient themselves in a certain fashion. Therefore, fungicidal activity can also be attributed to the greater number of hydrogen bonds between the ligand and protein. The preferred docked orientation obtained from PyMol shows the involvement of phenyl ring among the complex’s cystathionine–β-lyase+pyrimethanil; cytochromec- oxisdase+mancozeb; cch1+chitosan; peroxidase+probenazole; polyphenoloxidase+probenazole and Monooxygenase+mancozeb. The interactions with active site residues coupled with favorable binding energy proclaim that these compounds may serve as an effective surrogate for the fungicidal activity for respective fungal diseases undertaken in this study.
| Protein molecule | Energy | Bond energy | Angle | Torsion | Improper | Non-bond | Electrostatic constraint |
Total |
|---|---|---|---|---|---|---|---|---|
| cch1 | computed total | 99999900 | 47865.751 | 6530.439 | 11270.963 | 99999900 | -25291.15 | 19984572 |
| minimized | 99999900 | 57373.512 | 6878.568 | 13514.231 | 99999900 | -22681.95 | -1054117 | |
| cytochrome c oxidase. | computed total | 1175.463 | 1714.463 | 1234.89 | 614.488 | 2556.57 | -4988.83 | 2906.709 |
| minimized | 457.569 | 1051.842 | 1348.383 | 452.036 | -3136.71 | -5743.36 | -5561.242 | |
| monooxygenase | computed total | 150.699 | 1052.197 | 1372.626 | 429.567 | -4581.09 | -6028.68 | -7604.674 |
| minimized | 150.968 | 1052.756 | 1372.273 | 429.224 | -4583.22 | -6029.55 | -7607.55 | |
| cystathionine-β- lyase | computed total | 154.006 | 1052.666 | 1369.831 | 436.983 | -4314.39 | -5952.2 | -7253.106 |
| minimized | 150.699 | 1052.197 | 1372.626 | 129.567 | -4581.09 | -6028.68 | -7604.674 | |
| Peroxidase | computed total | 1979.069 | 2564.639 | 1338.56 | 1386.381 | 5130.31 | -7366.95 | 5031.997 |
| minimized | 577.743 | 1390.269 | 1382.799 | 525.87 | -646.28 | -8516.41 | -11096.001 | |
| Demethylase | computed total | 10054.204 | 12698.009 | 5917.408 | 4064.339 | 40620.33 | -15933.87 | 54720.422 |
| minimized | 3114.319 | 10336.391 | 6499.967 | 2789.294 | 3384.16 | -18825.32 | -7298.813 | |
| Polyphenoloxidase | computed total | 2537.4799 | 3019.232 | 1923.335 | 706.161 | -3085.18 | -9377.37 | -4267.331 |
| minimized | 684.619 | 1865.642 | 1854.55 | 520.567 | -9780.65 | -10855.62 | -15710897 | |
| pdb2c7y | computed total | 1949.017 | 3843.939 | 4559.577 | 537.308 | -23915.9 | -16057 | -29083.145 |
| minimized | 725.099 | 2646.74 | 4113.555 | 659.865 | -26382.14 | -18227.13 | -36464 | |
| pdb1z92 | computed total | 668.557 | 1449.779 | 1585.445 | 259.907 | -4613.03 | -6483.19 | -7132.528 |
| minimized | 189.313 | 949.378 | 1593.928 | 274.766 | -7227.92 | -7205.94 | -11426.379 | |
| 4ybn | computed total | 2790 | 2612 | 2339 | 527.906 | -10786.92 | -8728 | -11185.759 |
| minimized | 434.41 | 1415.004 | 2147.156 | 383.284 | -12801.47 | -9618.82 | -18040.439 | |
| 1llw | computed total | 10646.304 | 11601.042 | 13951.29 | 27010.392 | 42116.07 | -33670 | 47345.391 |
| minimized | 1496.51 | 8873.74 | 12787.144 | 2362.402 | -38122.54 | -36370.85 | -49310.594 | |
| 1bcc | computed total | 6106.357 | 13449.307 | 8779.513 | 199.139 | 455589.78 | -52798.91 | 433118.118 |
| minimized | 1549.8111 | 8960.406 | 9018.808 | 1997.091 | -42361.42 | -55373.28 | -76208 | |
| 1wyg | computed total | 2790.378 | 6780.973 | 5982.44 | 1162.124 | -28140.39 | -30146.8 | -41571.273 |
| minimized | 1045.72 | 4580.17 | 5990.482 | 1270.283 | -4066.79 | -33601.16 | -61384.301 | |
| 2v4h | computed total | 2151.561 | 2338.347 | 3590.485 | 507.618 | -8823.95 | -12582.51 | -12818.444 |
| minimized | 469.993 | 1723.622 | 3255.906 | 591.262 | -13110.95 | -13996.41 | -211066.57 | |
| 2x3n | computed total | 839.117 | 1950.879 | 1780.14 | 365.728 | -11009.44 | -13771.89 | -19845.465 |
| minimized | 326.398 | 1137.571 | 1624.511 | 344.848 | -12225.47 | -14812.88 | -23605.021 | |
| 3e6g | computed total | 6312.943 | 9034.845 | 10726.083 | 1810.986 | -33945.41 | -25731.26 | -31791.814 |
| minimized | 1285.313 | 9605.19 | 6366.115 | 1836.115 | -45334.39 | -29137.6 | -55378.848 | |
| 4h33 | computed total | 346.377 | 762.248 | 491.11 | 57.169 | -2162.96 | -1485.56 | -1991.615 |
| minimized | 87.224 | 412.521 | 458.184 | 96.46 | -2646.66 | -1844.4 | -3436.671 | |
| 4hex | computed total | 981.508 | 1775.327 | 1759.954 | 304.914 | -6471.42 | -3502.69 | -5152.405 |
| minimized | 244.655 | 1306.058 | 1540.014 | 489.77 | -8745.18 | -5824.17 | -10988.855 | |
| 4je5 | computed total | 6908.802 | 12186.805 | 11406.286 | 2389.735 | -47460 | -45495.62 | -60064 |
| minimized | 1207.18 | 7813.069 | 10566.146 | 1863.295 | -69234.39 | -49919.98 | -97704.68 | |
| 403t | computed total | 2396.019 | 5633.731 | 5477.164 | 604.164 | -25639.02 | -17119.66 | -28647.691 |
| minimized | 539.266 | 2763.999 | 5059.61 | 587.125 | -26229.8 | -19202.6 | -36482.406 |
Table 3: Energy values as calculated for the reported and screened protein sequences.
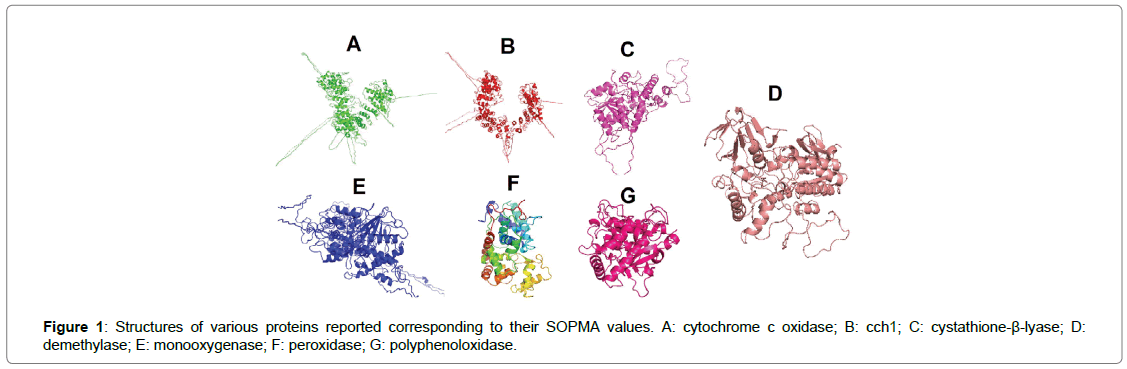
Figure 1: Structures of various proteins reported corresponding to their SOPMA values. A: cytochrome c oxidase; B: cch1; C: cystathione-β-lyase; D: demethylase; E: monooxygenase; F: peroxidase; G: polyphenoloxidase.
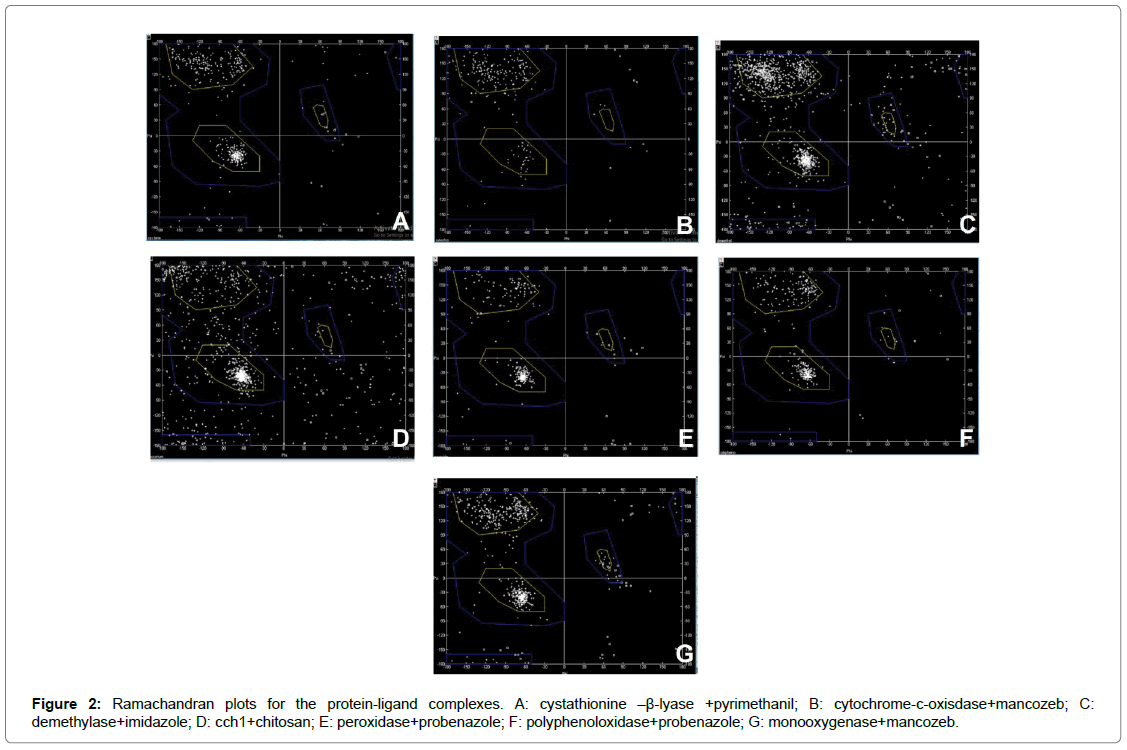
Figure 2: Ramachandran plots for the protein-ligand complexes. A: cystathionine –β-lyase +pyrimethanil; B: cytochrome-c-oxisdase+mancozeb; C: demethylase+imidazole; D: cch1+chitosan; E: peroxidase+probenazole; F: polyphenoloxidase+probenazole; G: monooxygenase+mancozeb.
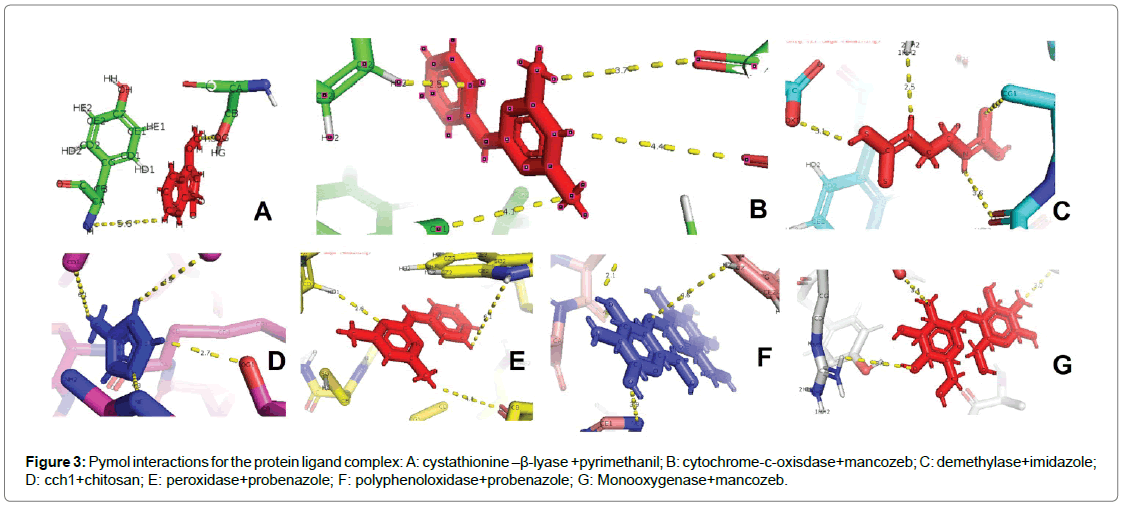
Figure 3: Pymol interactions for the protein ligand complex: A: cystathionine –β-lyase +pyrimethanil; B: cytochrome-c-oxisdase+mancozeb; C: demethylase+imidazole; D: cch1+chitosan; E: peroxidase+probenazole; F: polyphenoloxidase+probenazole; G: Monooxygenase+mancozeb.
| Complex name | Total amino acid in disallowed region | Glycine | Amino acid other than glycine |
|---|---|---|---|
| Cystathionin-β-lyase + Pyrimethanil | 24 | 8 | 16 |
| Cytochrome c oxidase + Mancozeb | 12 | 8 | 4 |
| Demetylase + Imidazole | 67 | 24 | 43 |
| cch1 + Chitosan | 126 | 20 | 106 |
| Peroxidase + Probenazole | 13 | 8 | 5 |
| Polyphenoloxidase + probenazole | 10 | 4 | 6 |
| Monooxygenase + Mancozeb | 28 | 18 | 10 |
Table 4: R-plot values for various protein ligand complexes under study.
| Protein molecule+ligand | Energy | Bond energy | Angle | Torsion | Improper | Non-bond | E constraint | Total |
|---|---|---|---|---|---|---|---|---|
| cystathionine beta lyase +pyrimethanil | computed | 754.168 | 2645 | 2124.87 | 855.516 | -6693.7 | -8790.4 | -9104.5 |
| minimized | 364.097 | 2211 | 2220.61 | 822.359 | -9506.8 | -9655.8 | -13544 | |
| cytochrome c oxidase + mancozeb | computed | 411.494 | 1328 | 1345.35 | 465.451 | -3044.6 | -5488 | -4981.9 |
| minimized | 410.283 | 1330 | 1344.52 | 464.577 | -3050.2 | -5494.2 | -4994.8 | |
| cch1+chitosan | computed | 1E+08 | 47865.7 | 6532.96 | 11271 | 1E+09 | -25298 | ####### |
| minimized | 1E+08 | 57299.1 | 6882.43 | 13519.3 | 1E+08 | -22703 | -105307 | |
| monoxygenase+mancozeb | computed | 884.427 | 4385 | 2793.63 | 1575.53 | -6411.3 | -13822 | -10595 |
| minimized | 1190.36 | 3446 | 2978.15 | 1339.49 | -11408 | -15038 | -17492 | |
| demethylase+ imidazole | computed | 2877.41 | 11201.5 | 6468.63 | 3444.65 | 4289.96 | -17828 | 10454.3 |
| minimized | 2346.93 | 9941.15 | 7095.02 | 3006.76 | -10658 | -20348 | -8616.9 | |
| peroxidase+probenazole | computed | 549.414 | 1684 | 1378.26 | 696.008 | -6387.6 | -8097.1 | -10177 |
| minimized | 299.184 | 1395 | 1417.69 | 628.369 | -7776.2 | -8738.9 | -12775 | |
| polyphenoloxidase+ probenazole | computed | 632.708 | 2480 | 1847.79 | 581.919 | -9627.8 | -10483 | -14568 |
| minimized | 353.26 | 1874 | 1897.32 | 523.199 | -10921 | -11145 | -17455 |
Table 5: Energy estimation values for the protein ligand complex.
Various molecules were taken under the study, for being the target ligands to bring about a fungicidal reaction in the plant pathogen system. Ligands identified through this study allow us to make plant host fight against the fungal pathogen and prevent the occurrence of the disease. The interactions have been thoroughly studied with various softwares for the overall evaluation of the drug molecule designed and to study its overall effects for the overall higher efficacy and to prevent the occurrence of the fungal disease and management of the fungal pathogens in agriculture against various economically valuable plants.
Citation: Mishra P, Eswaran M, Raman NM, Kaul T (2019) Probing of Phytofungal Proteins for Fungicidal Activity by Molecular Docking. J Proteomics Bioinform 12: 079-084. doi: 10.35248/0974-276X.19.12.499
Received: 15-Mar-2019 Accepted: 07-May-2019 Published: 14-May-2019 , DOI: 10.35248/0974-276X.19.12.499
Copyright: © 2019 Mishra P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.