Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2008) Volume 1, Issue 9
Constitutively phosphorylated proteins in Human fetal liver (HFL) aged 16-24 wk of gestation were studied using a 2-DE step followed by western blotting detection and MS identification. We found 166 phosphorylated protein spots with quantitative information and identified 101 gene products. Of theses identified proteins, 57 contain phosphoserine, 49 contain phosphothreonine, 51 contain phosphotyrosine, and 64 were newly identified phosphorylated proteins. The possible phosphorylation sites were further predicted using Netphos, ScanProsite and Scansite programs and most proteins were predicted the same site by at least 2 programs. Integrating the functional categories, protein abundance and the degree of phosphorylation of these proteins, we found proteins related to carbohydrate, lipid and amino acid metabolism were highly expressed with also the high degree of all serine, threonine and tyrosine phosphorylation; proteins associated with hematopoiesis were relatively highly expressed but with a relatively low degree of phosphorylation at serine, threonine and tyrosine; the proteins for signal transduction; biosynthesis of secondary metabolites and those whose function were unknown were lowly expressed, but with the zhigh degree of phosphorylation and interestingly, serine was the main phosphorylated amino acid of signal transducers; threonine in enzymes of biosynthesis of secondary metabolites; and tyrosine in proteins with unknown function.
Keywords: Phosphorylated proteins; Human fetal liver; Mass spectrometry
2-DE: Two-dimensional gel Electrophoresis; CAMP: cAMP- and cGMP-dependent protein kinase; CDK: Cyclin Dependent Kinase; CK2: Casein Kinase II; GAPDH: Glyceraldehyde- 3-phosphate Dehydrogenase; HFL, human fetal liver; hnRNP K, heterogeneous nuclear ribonucleoprotein K; MALDI-TOF MS, matrix assisted laser esorption/ionization-time of flight mass spectrometry;
PMF, peptide mass fingerprinting; PKC, protein kinase C; PTM, post-translational modifications; PVPD, polyvinylpyrrolidone; Q-TOF MS: Quadrupole-time of Flight Mass Spectrometry; TCA: Trichlorcacetic Acid.
A wide variety of post translational modification (PTM), such as phosphorylation, glycosylation, methylation, and acetylation, are known to play key roles in many cellular processes. Protein phosphorylation is one of the most biologically important PTM. The reversible phosphorylation of proteins is central to the regulation of most aspects of cell function such as cell cycle, development and differentiation, metabolism, nerve activity, muscle constriction, transcriptional regulation and disease. Especially in signal transduction, protein phosphorylation is a key event. It is estimated that approximately one-third of all proteins in eukaryotic cells are phosphorylated at any one time (Zolnierowicz and Bollen, 2000) and that approximately 2% of the human genome codes for kinases and phosphatases (there are ~500 kinases and 100 phosphatases in humans). (Venter et al., 2001) The most common and important sites of phosphorylation in eukaryotes occur on serins, threonine and tyrosine residues (Yan et al., 1998). Eukaryotic protein phosphorylation is involved in many fundamentally physiological processes and abnormal phosphorylation is now recognized as a cause or consequence of many human diseases such as cystic fibrosis, Alzheimer’s disease and severe combined immunodeficiency. While many phosphoproteins have been identified, the gene products from the estimated 30,000 genes, will certainly add to the number of sequences encoding for proteins whose primary regulation occurs via phosphorylation (Cohen, 2002). To analyze this additional layer of protein diversity and to reveal its complexity, the traditional protein- by-protein approach clearly will not suffice to meet the needs of deciphering the phosphoproteome. Therefore, proteome-based technology and bioinformation is required to analyze phosphorylation of proteins.
The liver is the largest organ in the human body, probably the second only to the brain in organ complexity; displays the main digestive function for the metabolism of most substances such as carbohydrates, fats, proteins, vitamins, and hormones. In addition to digestion, it functions in the biodegradation of xenobiotics, production of various plasma proteins and production of red blood cells during embryonic development. Human fetal liver aged 16-24 wk of gestation is a major stage of fetal hematopoiesis in man, and is at the critical turning point between immigration and emigration of the hematopoietic system. It simultaneously consists of hepatic parenchymal cells and hematopoietic stem/progenitor cells. Therefore, the unique characteristics of the fetal liver at this stage are worthy of investigation. Since phosphorylation of proteins plays a pivotal role in biological processes, what we intriguingly want to know is the variety and the amount of constitutively phosphorylated proteins in the HFL and what kinds of biological processes they involved in. It is obvious that the establishment of a detailed catalog of phosphorylated proteins in HFL, the discovery of novel phosphorylated proteins from it will certainly facilitate our understanding of the mechanisms of coexistence of hepatic and hematopoietic systems in fetal liver.
Our laboratory have not only sequenced 13,077 expressed sequence tags (ESTs) from a cDNA library of HFL22w and generated a gene expression profile including 1,660 genes (Yu et al., 2001), but established the protein expression profile of HFL, which was composed of at least 2,495 distinct proteins (Ying et al., 2006).By analyzing the compiled expression profiles of liver at different developmental stages, we found some tissue-specific and developmental-stage specific gene groups that are likely to play important roles in some definite functional features. The phosphorylation state of a protein can not generally be directly controlled by gene-expression and should thus be determined at the protein level. In the current investigation, we have established a constitutively phosphorylated proteins profile of HFL aged 16-24 wk of gestation. Identifications of phosphorylated proteins in HFL were performed by proteomics combined with 2-DE, western analysis using anti-phosphoserine, antiphosphothreonine, anti-phosphotyrosine antibodys, and mass spectrometry. Computer assisted predictions of phosphorylation sites and protein kinases possibly involved have been done to understand the large size of information. As a result, we found protein phosphorylation extensively involved in the important biological function of HFL and these constitutively phosphorylated proteins has unique functional characteristics in HFL aged 16-24 wk of gestation.
Experimental Procedures
Antibodies
Phosphoserine Detection Kit (mouse mAb, Calbiochem); Phosphothreonine Detection Kit (mouse mAb, Calbiochem); Phosphotyrosine Ab-1(PY20, mouse mAb, NeoMarkers); goat-anti-mouse IgG and ECL luminescence kit were purchased from Santa Cruz Corporation.
Sample preparation
Chinese volunteers underwent induction of labor with water bag in Beijing Northern Taiping Road Hospital. Eight livers of Homo sapiens fetuses in gestation period of 16-22 weeks were used for proteomic analysis after obtaining informed consent. All procedures were conducted in accordance with protocols approved by the local institution’s ethical committee. Liver samples were immediately washed completely with iced PBS at 4°C. Liver tissue fragments (0.2g) were placed in mortar and liquid nitrogen was poured in it. Crush the fragments until it become powder-like substances. The powder was homogenized in 1mL hypotonic buffer (20mM Tris pH 7.5, 1.5mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton X-100, 2.5 mM Sodium pyrophosohate , 1mM -Glycerophosphate, 1mM Na3VO4 and proteinase inhibitor cocktail) in a Teflon glass homogeniser and centrifuged at 12,000 rpm for 30min at 4°C. The supernatant was precipitated using a solution of 20mM DTT, 10% TCA in acetone 1h at -20 °C and subsequent washing of pellets with 20mM DTT in cold acetone. Pellets are resuspended in 800μl lysis buffer: 7M urea, 2M thiourea, 4% CHAPS, 1% DTT, 2% amphalyte (pH3-10) and proteinase inhibitor cocktail. The sample was applied to the IPG strips. The protein content was determined by the Coomassie blue method (Bradford, 1976).
2D-gel Electrophoresis and Immunoblot Analysis
2-DE procedures were carried out as described previously (Wan et al., 2001) with little modifications. Samples of 0.5 mg total protein were applied on 7 cm immobiline DryStrips (pH4-7 and pH 6-11) with the Amersham IPGphor in combination with Bio-Rad mini 2D-gel 12% non-gradient SDS-PAGE. Before application, for pH4-7 samples were diluted to a total volume of 250 l with 8 M urea, 2% CHAPS, 0.5% IPG buffer (pH 4–7), 18 mM DTT and a trace of bromophenol blue; for pH 6-11 samples were diluted to a total volume of 250 μl with 7 M urea, 2M thiourea, 4% CHAPS, 10% isopropanol, 5% glycerol, 2% IPG buffer (pH 6–11), 2.5%DTT and a trace of bromophenol blue. The programmed condition of IEF in IPGphor for preparative 2-DE was as follows: 30 v, 6hr (step and hold); 60v, 7hr (step and hold); 200 v, 1hr (step and hold); 500 v, 1hr (step and hold); 1,000 v, 1hr (step and hold); 8,000 v, 1hr (gradient); 8,000 v, 9hr (step and hold) ; 8,000 v, 10hr (step and hold, for pH 6- 11 dryStrips only). Proteins from SDS-PAGE were stained with Coomassie Blue R250, or electroblotted onto a PVDF membrane (Amersham Pharmacia). For proteins with pI between pH4 and 7, the transfer buffer is composed of 25mM Tris, 192mM glycine and 20% methanol; for proteins with pI between pH6 and 11, the transfer buffer is 10 mM CAPS pH11.0. Membranes were blocked by 3% BSA (fraction V) and 3% PVPD in Tris-buffered saline (TBS; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl)–0.1% Tween 20 for overnight, incubated for 1.5 h with the anti-phosphoserine, anti-phosphothreonine, anti-phosphotyrosine antibodies respectively. Antibody concentration for anti-phosphoserine and anti-phosphothreonine was 1 μg/ml; antibody dilution for anti-phosphotyrosine antibody was 1:1,000. Secondary antibodies was goat-anti-mouse IgG (1:5,000–4,000) conjugated to horseradish peroxidase. Antibodies were diluted with dilution buffer (1% BSA [fraction V] and 1% PVPD in Tris-buffered saline [TBS; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl]–0.1% Tween 20 ). Antibody complexes were detected by chemiluminescence using the ECL kit and exposed to X-ray film (Kodak). All the experiments were repeated 3 times and the stable detection were analysed further.
Image Analysis
SDS-PAGE and film images were scanned by GS-710 calibrated imaging densitometer (Bio-Rad) and the semiquantitatively analyzed using PDQUEST 7.1.0 software package (Bio-Rad, USA). Intensity levels were normalized between gels and films by expressing the intensity of each spot in a gel or film as PPM (×1,000,000).
In-gel Tryptic Protein Digestion and Mass Spectrometric Analysis
Coomassie-stained protein spots were excised from 2- DE gel and transferred to a 96-well plate. The gel plugs were washed with 50% ACN/25mM NH4CO3 at least for half an hour and dehydrated with ACN. Then 5 μl trypsin solution (10 ng/μl in 25mM NH4HCO3) was added into each of the wells to reswell the gel plugs. After tryptic digestion, the peptides were extracted with 8 μl 0.1% TFA/50% acetic acid in an ultrasonic cleaner (KQ-250B, Kunshan Ultrasonic Instrument, Jiangsu, China) for 20 min.
One microliter of extracted peptides mixture solution was mixed with same volume of saturated a-cyano-4-hydroxytrans- cinnamic acid solution in 0.1% TFA, 50% ACN and dispensed on to 96-well target for MALDI-TOF mass spectrometer (MALDI-R, Waters) analysis. Spectrum acquisition was first calibrated with lock mass ACTH (adrenocorticotropic hormone fragments 18-39, MH+: 2465.199 Da) and then with auto-digested peak of porcine trypsin (MH+: 2211.105 Da). Mass accuracy was set at 50 ppm in peptide mass fingerprinting, and one possible missed cleavage for trypsin digestion was selected in MASCOT searching against SWISS-PROT and TrEMBL protein database; the proteins with a confidence level of more than 95% and that matched at least four peptides were considered to be a significant identification.
When the MALDI peptide mass maps failed to identify the protein unambiguously, CapLC-ESI Q-TOF MS was used to identify protein by peptide sequence tags in the protein sequence database. Nanoscale RP HPLC of the peptide mixture was carried out on a CapLC liquid chromatography system (Waters). Peptide mixtures were injected onto a precolumn (300μm -inner diameter×5-mm PepMap C18, 3-mm length; LC Packings, Amsterdam, The Netherlands) for desalting. The separation was performed on a capillary C18 column (75 μm×15 cm; LC Packings) by running a gradient of 4% B (80% ACN, 0.1% formic acid) to 50% B TOF mass spectrometer (Q-TOF Micro, Waters) at a flow rate of about 200 nl/min. The positive ion mode was used; the spray voltage was set at 3.2 kV, and the spray temperature was 80 °C. MS/MS spectra (maximum 7.7 s) were acquired from the four most intense ions in each full scan with dynamic exclusion within 55 s. Raw data were processed using MassLynx Version 4.0 (smooth 3/2 Savitzky Golay and center four channels/80% centroid), and the resulting MS/MS dataset was exported into the pkl files. The data was searched against the SWISS-PROT and TrEMBL protein database by MASCOT. Mass tolerance of peptide precursor and its daughter ions was set at 0.2 Da in peptide sequence tag, and one possible missed cleavage for trypsin digestion was selected. Protein identifications were performed based on probability-based Mowse scoring algorithm with a confidence level of 95%.
Functional Classification
The assignment of protein function is based on the known function according to current annotation of SWISS-PROT (http://www.expasy.ch), classifications provided in the KEGG database (http://www.genome.jp/kegg), GO annotation (http://www.geneontology.org) and related publications .
Prediction of Phosphorylation Motifs
The phosphorylation sites were predicted by NetPhos program from website (http://www.cbs.dtu.dk/services/ NetPhos), ScanProsite (http://www.expasy.ch/tools/ scanprosite/) and Scansite (http://scansite.mit.edu/). ScanProsite program can predict cAMP- and cGMP-dependent protein kinase (CAMP), protein kinase C (PKC) and casein kinase II (CK2) phosphorylation site for serine or threonine phosphorylation, tyrosine kinase (TYR) phosphorylation site for tyrosine phosphorylation. Protein kinase involved in phosphorylations and the possible binding proteins of phosphorylated proteins were predicted by Scansite program.
Identified proteins were quantified by tracking pairs of labeled and unlabeled peptides from the MS spectra, and it required at least a Leu-containing peptide to quantify. Protein abundance was calculated as ratios of the peak intensity of the fragment ions from the labeled versus the unlabeled peptides. Ratios were calculated from the average of all quantified peptides for a single protein.
Identification of Phosphorylated Proteins
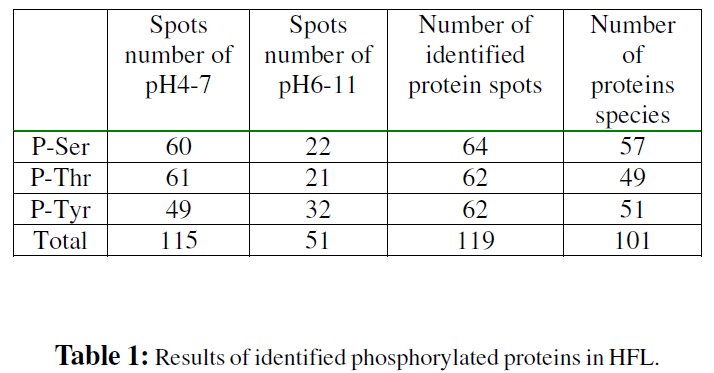
HFL proteins were separated by 2-DE in two pI ranges, 4-7 and 6-11. To observe as many proteins as possible, we did not apply protein prefractionation or immunoprecipitation prior electrophoresis. However, enrichment of the total protein amount by total protein precipitation was performed to detect proteins present in low amounts as described before(Rabilloud et al., 1997). The gels were visualized by Coomassie Blue staining, or blotted to PVDF membrane. Proteins containing phosphoserine, phosphothreonine and phosphotyrosine were identified by detection with antiphosphoserine, anti-phosphothreonine and antiphosphotyrosine monoclonal antibodys respectively and ECL chemiluminescence detection kit (Fig. 1). The positive and negative controls were used to monitor the specificity and selectivity of these antibodies, which showed that the antibodies and the WESTERN system we used is optimal (FigS1).The immunostained PVDF membrane were exposed to x-ray film. The film images were scanned using PDQUEST 7.1.0 software connected to GS-710 calibrated imaging densitometer. We found 166 phosphorylated protein spots in HFL: 115 spots in pI range of 4-7 and 51 spots in pI range of 6-11. Immunostained spots detected in any one of the immunoblot analyses were overlaid and depicted by arrows numbering in the gels pH4-7 (Fig. 1A) and pH6- 11 (Fig. 1E). The corresponding anti-phosphoserine immunostained gels were shown in Fig. 1B and Fig. 1F, anti-phosphothreonine immunostained gels were shown in Fig. 1C and Fig. 1G, anti-phosphotyrosine immunostained gels were shown in Fig. 1D and Fig. 1H. Protein spots detected on the immunoblot were cut out from the corresponding gel and subjected to in-gel digestion with trypsin. MALDI-TOF and LC-MS/MS were used to identify proteins. Low abundance proteins (weakly stained with Coomassie Blue) were identified by pooling spots from more than 3 gels.
Figure 1: 2D-gel images visualized by Coomassie blue staining and immunostaining. HFL proteins were analyzed by 2Dgel and visualized by staining. One of the best images was presented. (A) 2D Coomassie blue-stained gel map of pI range 4- 7. (B) 2D immuno-stained gel map of pI range 4-7 with anti-phosphoserine monoclonal antibody. (C) 2D immuno-stained gel map of pI range 4-7 with anti-phosphothreonine monoclonal antibody. (D) 2D immuno-stained gel map of pI range 4-7 with anti-phosphotyrosine monoclonal antibody. (E) 2D Coomassie blue-stained gel map of pI range 6-11. (F) 2D immuno-stained gel map of pI range 6-11 with anti-phosphoserine monoclonal antibody. (G) 2D immuno-stained gel map of pI range 6-11 with anti-phosphothreonine monoclonal antibody. (H) 2D immuno-stained gel map of pI range 6-11 with anti-phosphotyrosine monoclonal antibody. Numbered spots were excised and analyzed by in-gel trypsin digestion and MALDI-TOF MS or LCMS/ MS.
This allowed 166 phosphorylated protein spots to identify 140 proteins corresponding to 101 gene products. We observed at least 57 protein spots containing phosphoserine; 49 protein spots containing phosphothreonine and 51 protein spots containing phosphotyrosine. The detection results were summaried in Table 1 and Table S1. Of these, 25 proteins contain three kinds of phosphorylated amino acid residue, 35 proteins contain two of them. From certain spots, two or more proteins were identified (such as 20-1/2, 29-1/ 2, 30-1/2, 32-1/2, 41-1/2, 42-1/2, 49-1/2, 54-1/2/3/4, 69-1/2, 74-1/2, 78-1/2, 82-1/2, 92-1/2, 95-1/2, 109-1/2, 129-1/2), which were because both the very similar measured mass and the similar measured pI. Some different spots turned out to be the same proteins possibly due to modifications or degradation. A few proteins could not be identified because these were very weak spots which did not deliver a sufficient amount of peptides or peptide losses or because the MS data were insufficient for protein identification.
Phosphorylated Proteins Profile in HFL
On the basis of SWISS-PROT, KEGG database annotation, GO annotation and relative papers, the identified proteins can be subdivided into two sets: those can be assigned into known functional classes (97%) and those to which no functionality has been ascribed (3%). As expected, the phosphorylated proteins were participated in most physiological events of HFL. The distribution of the functionally classified proteins is shown in Figure 2 and the list of the 18 different functional categories is summarized in Table 2. However, this grouping is not exclusive because multi-functional genes belong to several classes.
Figure 2: Function distribution of the 166 phosphorylated proteins in HFL. Assignments were made on the basis of (i) information provided in the SWISS-PROT website (www.expasy.ch) (ii) classifications provided in the KEGG database (http://www.genome.ad.jp/kegg) (iii) known protein biological function.

Since phosphorylations may modulate protein activity and phosphorylation on specific amino acids can regulate different function of proteins, the protein level and the degree of 3 kinds of amino acid phosphorylation of each spot ranged on their functional categories was shown in Fig 3 and Fig S2. The quantity of each spot was normalized using PDQUEST 7.1.0 software package. From the figure we can see the phosphorylated proteins related to carbohydrate metabolism, lipid metabolism and amino acid metabolism were highly expressed with also the high degree of all serine, threonine and tyrosine phosphorylation. Secondly, phosphorylated proteins associated with hematopoiesis were relatively highly expressed but with a relatively low degree of phosphorylation at serine, threonine and tyrosine. Thirdly, the proteins for signal transduction; biosynthesis of secondary metabolites and those whose function are unknown were lowly expressed, but with the high degree of phosphorylation and interestingly, serine is the main phosphorylated amino acid of signal transducers; threonine in enzymes of biosynthesis of secondary metabolites; and tyrosine in proteins with unknown function.
Figure 3: Quantification of protein levels and the phosphorylation degree of three kinds of amino acids of each functional categories. The protein levels and the degree of phosphorylations of each spot was quantified and normalized using PDQUEST 7.1.0 software package and expressed in relative magnitude as a relative intensity. Red bars, protein; green bars, serine phosphorylation; yellow bars, threonine phosphorylation; blue bars, tyrosine phosphorylation. Functional category include: I Carbohydrate Metabolism; II Energy Metabolism; III Lipid Metabolism; IV Nucleotide Metabolism; V Amino Acid Metabolism; VI Metabolism of Cofactors and Vitamins; VII Biosynthesis of Secondary Metabolites; VIII Biodegradation of Xenobiotics; IX Folding, Sorting and Degradation; X Cell Growth and Death; XI Transcription and translation; XII Signal Transduction; XIII Hematopoiesis related; XIV Immune related; XV Transport activity; XVI Cell motility and structure; XVII Function unknown ; XVIII Unidentified.
We searched for what is known about the subcellular location of the species detected during the proteomic analysis. For 28% of the proteins no annotation existed in the SWISS-PROT database concerning their subcellular location. The annotated proteins were mainly localized in the cytosol and mitochondria (Fig. 4). The SWISS-PROT annotation about the subcellular location of each individual protein is given in Table 2.
Prediction of Possible Phosphorylation Sites of Each Protein
Large scale analysis of 101 phosphorylated proteins in HFL aged 16-24 wk of gestation was performed by computer assisted program, which allowed us to predict the possible phosphorylation sites. Prediction of phosphorylation sites were performed by there computer programs including NetPhos (http://www.cbs.dtu.dk/services/NetPhos/), ScanProsite (http://www.expasy.ch/tools/scanprosite/) and Scansite program (http://scansite.mit.edu/) shown in Table S2. All three programs predicted the same phosphorylation site in 40, 32 and 12 proteins for serine, threonine and tyrosine residule respectively; at least two of the three programs predicted the same phosphorylation site in 17, 17 and 36 proteins respectively.
Our initial goal was to gain a broad understanding of both the diversity and the abundance of constitutively phosphorylated proteins and the degree of phosphorylation in HFL aged 16-24 wk of gestation which has its tissue-specific and stage-specific functions. In fact, quantitatively study phosphorylated proteins in large scale in specific tissue or organ is still the challenge so far.
Here we have undertaken a proteomic approach using a 2-DE step followed by western blotting detection, MS identification and bioinformation to profile phosphorylated proteins in HFL aged 16-24 wk of gestation as one step of a long-term effort to explore the important function in this specific developmental stage of human fetal liver. Totally we found 166 phosphorylated protein spots and identified 140 proteins corresponding to 101 gene products. Of theses identified proteins, 37 (36.6%) proteins were previously identified phosphorylated proteins and 64 (63.4%) were newly identified phosphorylated proteins (Table 2). Then, we used computer assisted programs such as Netphos, ScanProsite and Scansite consecutively to validate the results. As the programs use unique algorithms for prediction of phosphorylation sites, combining and comparing the results from the three programs should give useful information (Kim et al., 2000). We observed at least 57 protein spots containing phosphoserine; 49 protein spots containing phosphothreonine and 51 protein spots containing phosphotyrosine. All three programs predicted the same phosphorylation site in 40, 32 and 12 proteins for serine, threonine and tyrosine residue respectively; two of the three programs predicted the same phosphorylation site in 17, 17 and 36 proteins respectively (Table 2). The results demonstrated that most phosphorylated proteins detected in HFL could be confirmed by at least two of the three programs. Although each program has different stringency, some predictions common to two of the programs were noted. The results are consistent with the previous findings for phosphorylation sites and corresponding kinases: transitional endoplasmic reticulum ATPase (spot 1) is phosphorylated on Ser-352, 746 and 748 (Klein et al., 2005); protein NDRG1 (spot 41-2) on Ser-330 (Olsen et al., 2006); Ser15, Ser78 and Ser82 in HSP beta-1 (spot 95-1) (Olsen et al., 2006; Umeda et al., 1997; Balamurugan et al., 1999; Beausoleil et al., 2004) glutamate dehydrogenase 1 (spot 122) on Ser-227(Villén et al., 2007); methylmalonate-semialdehyde dehydrogenase (spot 123) on Ser-490 (Olsen et al., 2006); hemoglobin beta chain (spot 149) on Ser-45 (Villén et al., 2007; Ballif et al., 2004); triosephosphate isomerase (spot 154) on Ser-21(Olsen et al., 2006); phosphatidylethanolamine-binding protein 1 (spot 159,160,163) on Ser-51(Olsen et al., 2006). Rat liver Sadenosylmethionine synthetasewas regulated by PKC phosphorylation on Thr-342 (Pajares et al., 1994), in human this site is Thr-341 (spot 51); mouse liver glycine amidinotransferase is phosphorylated on Thr-416, in human this site is Thr-417 (spot 54-2) (Villén et al., 2007); Thr-320 in PP-1 (spot 70) is phosphorylated by Cdc2 kinase (Kwon et al., 1997; Helps et al., 2000; Guo et al., 2002); annexin IV (spot 79) is phosphorylated on Thr-6 by PKC (Kaetzel et al., 2001); peroxiredoxin-1 (spot 161) on Thr-90 (Chang et al., 2002); peptidyl-prolyl cis-trans isomerase A (spot 164,165,166) on Thr-157 (Gevaert et al., 2005). Heterogeneous nuclear ribonucleoprotein H (spot 20-2) is phosphorylated on Tyr-306 (Rush et al., 2005); LIM and SH3 domain protein 1 (spot 90) on Tyr-171 (Tao et al., 2005); DJ- 1 protein (spot 115) on Tyr-67 (Rush et al., 2005). At least two of the three programs predicted the known sites correctly.
In the liver of a human fetus, besides the general metabolism of carbohydrates, fats and proteins, hematopoiesis which originated in the yolk sac, occurs in the liver from the 6th wk to the 7th month of gestation. After the immigration of the hematopoietic system into the fetal liver at 2 months of gestation, human fetal liver gradually becomes a major site of embryonic hematopoiesis, and, intriguingly, coexistence of hepatic and hematopoietic systems appears. The reversible phosphorylation of proteins is central to the regulation of most aspects of cell function. The portion of the phosphorylated proteins profile in any given cell type or tissue is not precisely known. A preliminary profile of phosphorylated proteins in this cell population was set up based on the analysis of 166 phosphorylated protein spots. Of these, 25 proteins contain three kinds of phosphorylated amino acid residue, 35 proteins contain two of them. Multisite phosphorylation can enable two or more biological activity modulations to operate in the same protein. It can also determine the extent and duration of a response and is the key to signal integration (Cohen, 2000). As expected, we identified phosphorylated proteins were participated in most physiological events of HFL, including tissue-specific and stagespecific functions of HFL aged 16-24 wk of gestation (such as biodegradation of xenobiotics account for about 3%, Cell Growth and Death 7%, and hematopoiesis related proteins 3%) and housekeeping proteins function (such as metabolism account for about 54% and proteins that function in transcription and translation 3%).
Although the phosphorylated proteins activities were not simply reflected by the abundance of phosphorylated proteins and the degree of phosphorylation, phosphorylated proteins profiling leads to the best approximation about them. Because the phosphorylated proteins profile could be analyzed in terms of both patterns and levels. The profile dramatically reflected the phosphorylated proteins in hepatic, hematopoietic and housekeeping activities of HFL as described above. The quantitative data should help us understand their functional distribution feature. First, phosphorylated proteins related to carbohydrate metabolism, lipid metabolism and amino acid metabolism were highly expressed with also the high degree of all serine, threonine and tyrosine phosphorylation. Since these phosphorylated proteins can modulate cell general metabolism and supply a large amount of material and energy to cell, they play an important regulation role in cell proliferation and differentiation of HFL in this developmental stage. Second, phosphorylated proteins associated with hematopoiesis (including Uroporphyrinogen decarboxylase, Delta-aminolevulinic acid dehydratase, Hemoglobin beta chain, Hemoglobin gamma- A and gamma-G chains) were relative highly expressed but the degree of protein phosphorylation was low. The relative high abundance of hematopoiesis related phosphorylated proteins indicated that HFL was a major site of embryonic hematopoiesis, which accordant with the genes expression profiling of HFL22w accomplished by our laboratory previously (Yu et al., 2001). Uroporphyrinogen decarboxylase and Delta-aminolevulinic acid dehydratase were invole the pathway of Porphyrin and heme biosynthesis, which play pivotal roles in the regulation of hematopoiesis (Abraham, 1991). Hemoglobin beta chain, Hemoglobin gamma-A and gamma-G chains are the composition of red blood cells. The appearance of adult-type hemoglobin (hemoglobin beta chain) in HFL aged 16-24wk of gestation was agree with the conclusion of Choi et al., (1995) that the transition of hemoglobin type from fetal to adult form has already begun in the 22-wk-old fetal liver before the bone marrow takes over the hematopoietic function (Choi et al., 1995). Since some relative highly expressed proteins of them such as hemoglobin gamma-G chains whose hosphorylated states were never discovered before, these proteins might be fetal liver specific phosphorylated proteins which can regulate the hematopoiesis in HFL with relative low degree phosphorylation. Third, the proteins for signal transduction; biosynthesis of secondary metabolites and those whose function are unknown were lowly expressed, but with the high degree of phosphorylation and interestingly, serine is the main phosphorylated amino acid of signal transducers; threonine in biosynthesis of secondary metabolites; and tyrosine in proteins with unknown function. These low abundance proteins with high degree phosphorylation may play pivotal role in HFL. The unique regulation function of specific phosphorylated amino acid in HFL needs study further. Therefore the profile reflected the unique functional characteristics of phosphorylated proteins in HFL of this developmental stage. The liver development during various stages was apparently under the control of sequential phosphorylated proteins expression as the dominant, though perhaps not exclusive, mechanism. Therefore, in future using this proteomics approach to profile phosphorylated proteins of different development stage in human liver will did help us understand more about the functional features of HFL and identify specific phosphorylated proteins playing important roles during human liver development.
In summary, proteomic analysis combined with 2D-gel and western blotting, and mass spectrometry is a powerful tool for globally identifying the phosphorylated proteins in HFL. This is the first comprehensive study phosphorylated proteins in specific tissues by combining high throughput proteomic analysis and computer assisted methodology. However, to systematically characterize these phosphorylated proteins involved in the molecular mechanism of fetal liver development and embryonic hematopoiesis, several approaches, such as microarray and yeast two-hybrid system technologies, should be used in grouping analysis of phosphorylated proteins expression kinetics and protein interaction in human fetal liver. The phosphorylation sites identification in high throughput by high sensitivity and precision MS such as TQ-FT is also a new direction in this area.
This work was partially supported by Chinese State Key Projects for Basic Research (973) (Nos. 2001CB510204, 2006CB910401, 2006CB910801 and 2006CB910602), Chinese State High-tech Program (863) (2006AA02A308), National Natural Science Foundation of China (30700356), National Natural Science Foundation of China for Creative Research Groups (30621063) and by the Beijing Municipal Key Project (H030230280290).
The authors declare that they have no competing financial interests.