Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2009) Volume 2, Issue 2
Selaginella bryopteris (L.) Bak is a resurrection plant. Uniquely, its detached fronds have the ability to survive desiccation similar to that of the whole plant. In order to understand the mechanisms of desiccation tolerance, proteome studies were carried out in this plant using detached fronds to reveal proteins that were differentially expressed in response to dehydration and rehydration. There was not much difference in electrolyte leakage between control, dehydrated and rehydrated fronds. During dehydration the plants showed only respiration and a drop in Fv/Fm values. Both fluorescence and photosynthesis regained totally after rehydration. About 250 protein spots were reproducibly detected and analyzed. From the putatively identified spots (proteins), it was observed that proteins involved in transport, targeting and degradation were expressed more in the desiccated fronds. These findings tentatively indicate that some of the proteins could contribute a physiological advantage to S. bryopteris under desiccation.
Keywords: Selaginella bryopteris, Desiccation tolerance, Fluorescence, Two-dimensional electrophoresis.
Plants have evolved a wide spectrum of adaptations to cope with the challenges of environmental stress. Of the various stresses, one major factor that limits the productive potential of higher plants is the availability of water. The International Water Management Institute (IWMI), a CGIAR Institute with headquarters in Sri Lanka, predicts that by year 2025, one-third of the world’s population will live in regions that will experience severe water scarcity (http://www.iwmi.cgiar.org/). This will surely impact on agriculture in these regions. Therefore, it has become imperative for plant biologists to understand the mechanisms by which plants can adapt to water deficit while retaining their capacity to serve as sources of food and other raw materials. Water deficit can affect plants in different ways. A mild water deficit leads to small changes in the water status of plants, and plants cope by reducing water loss and/or by increasing water uptake (Bray, 1997). The most severe form of water deficit is desiccation — when most of the protoplasmic water is lost and only a very small amount of tightly bound water remains in the cell. Tolerance or resistance to this severe water deficit determines productivity of the plants, especially as food sources.
An important contribution to our understanding of the mechanism of desiccation tolerance is derived from ‘resurrection plants’, which can survive even with <5% of their total water in the vegetative tissues and are able to regain normal metabolism and growth within several hours of rewatering.
(Ramanjulu and Bartels, 2002). Selaginella bryopteris (L.) Bak is one such resurrection plant, with an unique feature of detached fronds possessing a similar level of desiccation tolerance as that of whole plants. This was ascertained by studying various physiological parameters in intact plants as well as in detached fronds (data not shown). The desiccation and rehydration of detached fronds avoid interference from developmental regulation and long-distance signalling from other organs (Jiang et al., 2007). We intend to use it as a model system to gain a system-level understanding of responses to desiccation using multiple platforms that provide information about global transcript levels, proteomes, a wide range of metabolites, enzyme activities, and growth parameters.
Stress responses in plants cause changes in the structure and activity of one or more proteins. Therefore, characterizing these proteins and understanding their function is an important method for studying responses of the plants to given stress factors. Identifying and characterizing individual proteins is an enormous task that is greatly facilitated in the modern times by combining 2D-gel electrophoresis with mass spectrometry. Such a combination is a powerful tool for identifying large numbers of proteins. These techniques, in combination with the constantly expanding genomic and EST databases, also enable the simultaneous identification of a these proteins. These methods can therefore, be collectively called as the Proteome analysis methods.
Here, we report a proteome analysis of the changes in proteins that occur in the detached fronds of S. bryopteris, first when they are dehydrated and subsequently after they are rehydrated. Our study describes the putative identification of 9 dehydration-responsive proteins by mass spectrometry and further shows that proteins involved in transport, targeting and degradation were expressed more in the desiccated fronds than that in the original excised fronds or the re-watered fronds.
Plants of S. bryopteris were collected from wild (Mirzapur district, U.P., India; latitude 23°52´-25°32´ N & longitude 82°7´-83°33´ E) and maintained in the fern house of the Institute.
Fronds with three different water status were used in the study: Control fronds (C - these were freshly excised fronds with RWC 100%), dehydrated (S1-RWC 10%) and rehydrated(S2 - RWC 100-104%). In order to dehydrate the fronds, freshly detached fronds from well-hydrated plants were placed in petri plates and subjected to dehydration in dark at 25 °C (60% relative humidity) in a growth chamber. Control samples of detached fronds were kept fully hydrated under the same condition. Rehydration was achieved by keeping the dehydrated fronds on wet filter paper for 12 h
in dark.
Photosynthesis Parameters and Chlorophyll Fluorescence
Photosynthesis parameters and chlorophyll fluorescence were measured and recorded using LiCOR 6400 and PAM 2000 (WALZ) systems, respectively according to the manufacturer’s procedures. Five independent replicates were used for these physiological parameters.
Fronds were frozen using liquid N2, ground in frozen state in a chilled pestle and mortar to a fine powder. This powder was extracted with 50 mM Tris-HCl, pH 8.0, 25 mM EDTA, 500 mM thiourea and 0.5% 2-mercaptoethanol (BME). The extract was mixed with 10% cold TCA and 0.07% BME, and left overnight at -20 °C. The mixture was centrifuged at 4500 rpm for 10 min and the pellet was washed three times with acetone containing 0.07% BME. The pellet was then vacuum dried, solubilised in 0.1 M Tris-HCl, pH 8.0, 50 mM EDTA and 2% BME. Proteins were extracted with 2.5 ml Tris-buffered phenol and centrifuged at 4500 rpm for 10 min. After centrifugation, lower phenol phase was collected with the help of Pasteur pipette. To this 10 ml 0.1 M ammonium acetate in methanol was added and left overnight at -20 °C. The mixture was centrifuged at 4500 rpm for 10 min and pellet was dissolved in 0.1 M ammonium acetate in methanol and 1% BME. It was centrifuged at 6000 rpm for 10 min. It was washed twice with cold acetone. Pellet was dried and stored at -80 °C until further use. Total protein content was analyzed using the protein assay dye reagent (Bio-Rad).
Isoelectric focussing (IEF) and Polyacrylamide gel electrophoresis (PAGE) Fifty μg protein was used for Isoelectric focussing (IEF) with 7 cm IPG strips, pH 4 to 7 in Ettan IPGphor unit (Amersham-GE Healthcare). The IPG strips were rehydrated overnight with total protein diluted in 8 M urea, 2% CHAPS (w/v), 0.5% IPG buffer pH 4 to 7, 25 mM DTT, 0.001% bromophenol blue up to a volume of 135μl. After rehydration, focussing was done on Ettan IPGphor under following conditions: 200 V for 20 min, 450 V for 15 min, 750 V for 15 min, and 2000 V for 4 h for a total of 10 kVh. Then strips were equilibrated in a buffer containing 50 mM Tris-HCl, pH 8.8, 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 1% (w/v) DTT for 15 min, and another 15 min in the same buffer but with 2.5% (w/v) iodoacetamide replacing DTT. The second dimension was run in Hoefer mini-gel apparatus using 7 x 8 cm homogeneous 12% SDSPAGE gels. Electrophoresis was performed in a standard Tris-Glycine running buffer at a constant voltage of 200 V. Gels were silver stained and gel images were acquired with the BioRad Fluor-S Imager. The data was analyzed using ImageMaster 2D Platinum 5.0 software (Amersham Bioscience). Relative volume (% volume) was used to quantify and compare the spots. The protein expression patterns were determined as up-regulated (% volume increased by two-fold or more), down-regulated (% volume decreased by two-fold or more) and unchanged (% volume varied less than or upto two-fold). Three independent replicates were used for the proteomic analyses. Only those spots present in at least three gels of independent sets were included in the analysis.
Tryptic digestion of the protein spots excised from the gels, and sample preparation were performed according to Koistinen et al., (2002). Briefly, gel pieces corresponding to selected spots were destained and dehydrated by washing three times with 25 mM ammonium bicarbonate containing
50% acetonitrile. Destained particles were dried in a vacuum centrifuge concentrator and rehydrated in equal volumes of 0.1 μg μl-1 trypsin (Sigma) and 50 mM ammonium bicarbonate and samples were digested overnight at 37°C. Peptides were extracted twice with 50% acetonitrile containing 5% Tri-fluoroacetic acid. Gel particles were rehydrated with water, and two more extractions were performed with 50% acetonitrile containing 5% Trifluoroacetic acid. The recovered peptides were
concentrated to a final volume of 20 μl.
The tryptic peptides were analyzed using Thermo Finnigan LCQ Advantage max ion trap mass spectrometer having Finnigan Surveyor HPLC system connected to it. The 2 μl sample was introduced into the ESI source through Finnigan Surveyor autosampler. The column was Thermo Bio-Basic 100 X 1, 5 μM and the peptides were eluted with linear 25 min gradients of 5–95% (v/v) acetonitrile with 0.1% (v/v) formic acid in water at a flow rate of 40 μl min-1. The mass spectra were scanned in the peptide mass range of 300- 1800 Da and the maximum ion injection time was set at 50 nanoseconds. Ion spray voltage was set at 5.3 KV and capillary voltage 30.5 V. The MS scan was continued up to 20 min for recording final data. The MS/MS data were processed using BIOWORKS 3.1 SR1 and searched against NCBI databases (nr Protein sequences) with the MS/MS ion searching program MASCOT (http://www.matrixscience.com) and homologies detected were scored as Ion Score (Score = –10 * log P, where P is the probability that the observed match is a random event). For the identification of proteins, Mascot search parameters were set as follows: taxonomy, Arabidopsis thaliana; fixed modification, carbamidomethylation at cysteine; variable modification, oxidation at methionine, precursor mass tolerance 1.5 Da, fragmented mass tolerance, 0.5 Da, digestion enzyme, trypsin; allowed miss cleavage, 2. Default settings were used for other parameters.
Detached fronds from fully hydrated S. bryopteris were subjected to dehydration and rehydration as described in material and methods. The RWC of detached fronds decreased rapidly from 100% (control) to a stable 10% after only 6 hrs. Dehydrated fronds showed intense inward curling (Fig 1). During rehydration, a RWC of 104% was achieved after 12 hrs and fronds regained broadly the original morphology. Leaf folding during drying of plants has been proposed to prevent light-chlorophyll interaction and light-induced damage (Farrant and Sherwin, 1998). Electrolyte leakage is used to test the integrity of cell during dehydration and rehydration. There was not much difference in electrolyte leakage between control, dehydrated and rehydrated fronds (Fig 2), indicating that S. bryopteris had a fundamental mechanism to survive desiccation. Farrant et al., (1999) also reported similar findings with desiccation tolerant angiosperm Craterostigma wilmsii. Since the detached fronds of Selaginella also showed similar response to that of the whole desiccation-tolerant plant, C. wilmsii, our results suggest that detached fronds of S. bryopteris plant thus represents a simplified system to investigate the basis of desiccation tolerance especially by taking advantage of avoidance of possible developmental regulation and long-distance signaling from other organs.
Figure 1: Excised fronds of Selaginella bryopteris. As indicated in the figure, the control frond is a freshly excised frond, still in a fully hydrated state (RWC 100%) while the Dehydrated (RWC 10%) frond is one subjected to dehydration as explained in the materials and methods. Note the severe inward curling of the frond in dehydrated state. The dehydrated frond when re-watered as described in the materials and methods restores to a Rehydrated state (RWC 100-104%).
The fronds immediately after detachment and in a stillhydrated state showed Fv/Fm ratios around 0.8 indicating the functional photosystems (Fig 3). After dehydration the fronds showed a decrease in both net respiration and Fv/Fm ratios. Both fluorescence and photosynthesis regained totally after rehydration. This clearly showed that detached, desiccated S. bryopteris fronds fully revived their metabolism after re-hydration. In general, water deficit causes a reduction in the photosynthesis rate, resulting in a decline in the photochemical efficiency of PSII and electron transport rate in both, desiccation-tolerant as well as desiccation-sensitive plants (Ekmekci et al., 2005). The decline in PSII activity could represent a protective mechanism from toxic oxygen production in order to maintain membrane integrity and to ensure protoplast survival (Di Blasi et al., 1998). However, only proteins within the thylakoid membranes of resurrection plants remain stable during desiccation and rehydration (Schneider et al., 1993), whereas those of desiccation- sensitive plants are completely destroyed even after a short-term desiccation event (Deng et al., 2003).
About 250 protein spots were reproducibly detected and analyzed (Fig 4). Forty eight spots amongst these showed significant and reproducible changes in abundance. Twenty one of them were up-regulated and 27 were down-regulated. Among the differentially accumulated proteins, 30 with relatively greater changes in abundance were analyzed by LC MS/MS. However, peptides from only nine of these could be identified with high probability. Of the various proteins detected, the putatively identified proteins are listed in Table 1. Proteins involved in transport, targeting and degradation were more expressed in the desiccated fronds (Table 1; Fig 5). One such protein was putatively identified as Fbox/ LRR-repeat protein. The F-box/LRR repeat is a conserved domain that is present in large number of proteins with a bipartite structure. Through the F-box, these proteins
are linked to the Skp 1 protein and the core of SCFs (Skp 1- cullin-F-box protein ligase) complexes. SCF complexes constitute a new class of E3 ligases. They function in combination with the E2 enzyme Cdc34 to ubiquitinate G 1 cyclins, Cdk inhibitors and many other proteins and to mark them for degradation. The physiological roles of proteolytic enzymes are diverse, as they are necessary both for processing proteins from an inactive to active states and for recycling redundant/damaged polypeptides (Schwechheimer and Schwager, 2004). It has been known that protein degradation via the ubiquitin–proteasome pathway plays a pivotal role in controlling cellular processes, such as cell cycle progression and transcriptional control in eukaryotic cells (Hershko and Ciechanover,1998). It is possible that induction of proteolytic enzymes, together with the upregulation of translation-related factors, may be related to the biosynthesis of novel proteins involved in the drought resistance mechanisms. Rivero et al., (2007) have, on the other hand, shown that suppression of drought induced senescence provided outstanding drought tolerance in transgenic tobacco plants. Two ribosomal proteins, 40S RPS27 and 60S RPL27, were up regulated under desiccation stress (Table 1, Fig 5). Vincent et al., (2007) have reported that in grapevine shoots, there is an increased abundance of RPL39 in response to drought. In yeast, this protein is a 60S ribosomal subunit implicated in translational accuracy (Dresios et al., 2000).
Figure 4: Comparison of 2D gel maps of proteins isolated from detached fronds of S. bryopteris during dehydration and rehydration. The pH range is indicated along the top of each gel, and the sizes of MW markers (kDa) are indicated down the left-hand side. U1 to U8 – up-regulated; D1 – down-regulated.
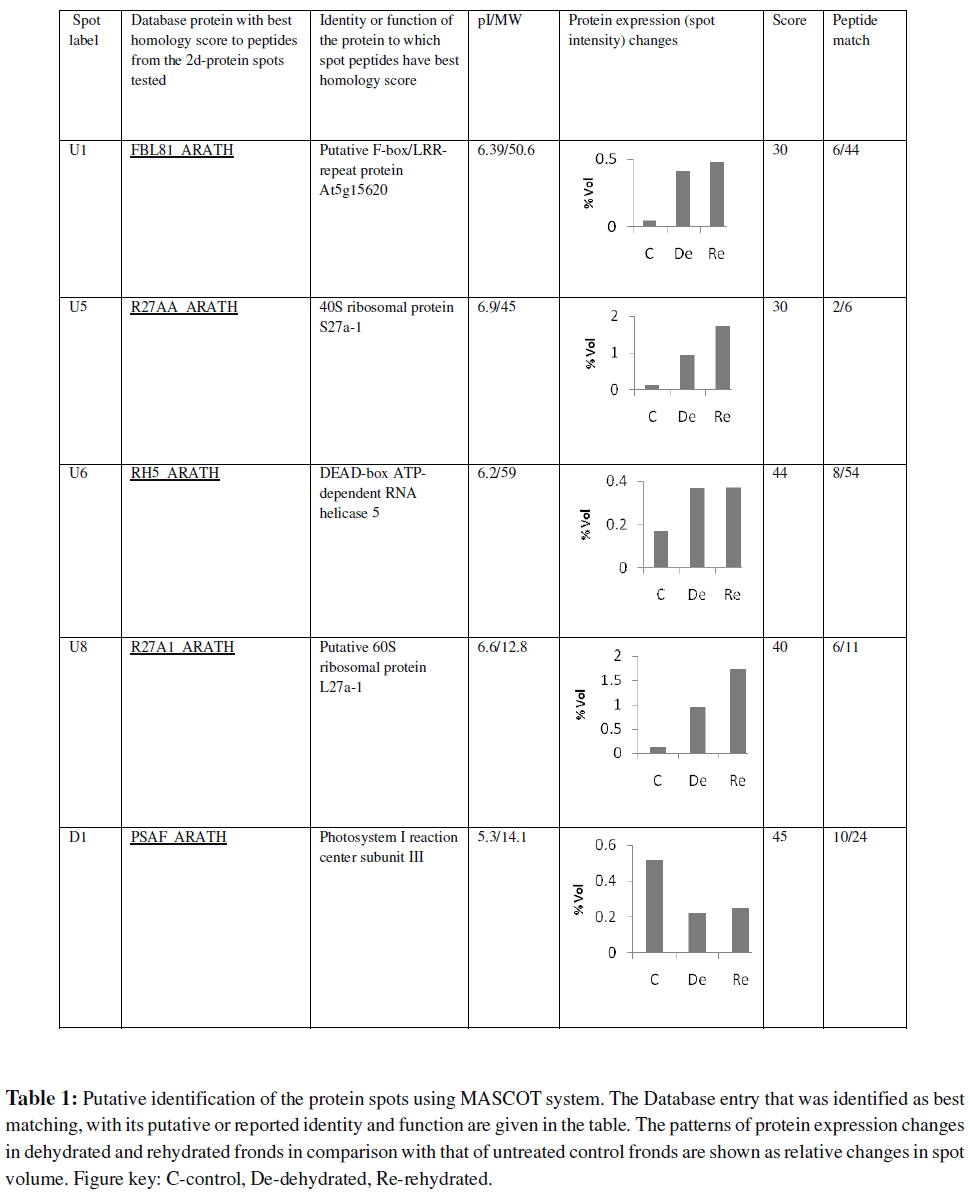
In the present study, a putative DEAD-box ATP-dependent RNA helicase 5 was also highly up-regulated in both dehydrated and rehydrated fronds (Table 1). DEAD-box RNA helicases have been implicated to have a function during stress adaptation processes, but their functional roles in plant stress responses remain to be clearly elucidated (Owttrim, 2006). Kim et al., (2008) found differential expression in transcript levels of two RNA helicases viz. AtRH9 and AtRH25 in Arabidopsis thaliana exposed to cold, drought or salt stress. A pea DEAD-box related helicase (PDH45) transcript was induced in pea seedlings in response to a range of abiotic stresses including salt (specifically Na+), dehydration, wounding and low temperature, leading to the suggestion that pdh45 transcript accumulates in response to general water stress caused by desiccation (Sanan-Mishra et al., 2005). Our results together with the above reports imply that DEAD-box RNA helicases may perform a crucial function directly involved in cellular responses to abiotic stress.
The only down-regulated protein that was putatively identified in our 2-D gels was a photosystem I reaction center subunit III homologue (Table 1, Fig. 5). This protein has been shown to participate in electron transfer from plastocyanin to P700. In the excised frond system this protein remained under-expressed even after rehydration, although photosynthesis was restored (Fig. 5). Sigfridsson and Oquist, (2006) showed that desiccation of tolerant species such as Cladonia impexa Harm and Trebouxia pyriformis Archibald causes a preferential energy distribution into photosystem I. These plants employ this strategy to avoid photodynamic destruction of the photosynthetic apparatus when photosynthesis is inhibited under dry conditions. The physical properties of the photosynthetic apparatus are thus of crucial importance in desiccation-tolerant plants. The photosynthetic
apparatus is very sensitive and liable to injury or even destruction during desiccation and needs to be maintained or quickly repaired upon rehydration (Godde, 1999). At peak stress intensity, the repression of genes encoding photosynthesis related proteins, Rubisco small subunit and PSI reaction center subunits VI and X, may also be due to severity of the stress and could indicate the beginning of senescence (Bogeat Triboulat et al., 2007). What influences the adaptation of the resurrection plant to withstand the stress and its severity is an important question, answers to which may have profound implications on stress related studies in crop plants with particular emphasis on their productivity in stressed conditions.
In summary, this paper has presented a preliminary study of the protein expression profile in response to dehydration and subsequent rehydration using the excised frond of the resurrection plant Selaginella bryopteris. The study showed the possible involvement of proteins involved in transport, targeting and degradation were expressed more in desiccated fronds and that all expression changes were not reversed when the desiccated fronds were rehydrated. Such changes in the fronds could contribute towards a physiological advantage for withstanding potential damage that can be caused by desiccation. However, identification of more protein spots as well as in planta studies using potted whole plants of Selaginella bryopteris may be required for a better understanding of not only desiccation stress but also possible restoration of function after the plants are destressed.
We thank Director, NBRI for his help and encouragement. This work was carried out under Supra Institutional Project (SIP-09) funded by Council of Scientific and Industrial Research, India.