Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2008) Volume 1, Issue 3
Development of strategies that allow for identification of secreted factors in conditioned media is of significance for a wide range of research areas. Secreted factors are involved in intercellular communication and might also be biomarkers of potential clinical importance, used for early detection and diagnosis of disease. The aim of this study was to investigate whether metabolic labeling combined with mass spectrometry could be used to identify secreted proteins in serum-containing conditioned medium. Earlier proteomic studies of conditioned media have been performed on cells cultured in serum-free media. In the present study the fact that only the proteins derived from the cells contain the incorporated isotopically labeled amino acid was taken advantage of, making it possible to differentiate released proteins from medium proteins. The second objective was to examine whether any quantitative differences in the secretion profiles between primary astrocytes and astrocytes in a scratch injury model of reactive astrogliosis could be found. To our knowledge, this is the first study to identify secreted proteins in serum-containing medium using a proteomic approach involving stable isotope labeling by amino acids in cell cultures and mass spectrometry.
Keywords: Proteomics, Mass spectrometry, Serum-containing conditioned medium, Secreted proteins, Astrocytes.
2D: Two-dimensional;
FT-ICR: Fourier Transform Ion Cyclotron Resonance;
LC-MS: Liquid Chromatography Mass Spectrometry;
LTQ: Linear Ion Trap; MS: Mass Spectrometry; SILAC: Stable Iisotope Labelling by Amino acids in cell Culture
Several studies have demonstrated that conditioned media contain secreted factors that have significant biological effects. Secreted factors have been proposed to be involved in intercellular communication. They might also be a source for early detection and diagnosis of disease, and identification of released factors, including proteins, might help to discover novel biomarkers of potential clinical significance. In fact, factors secreted by cancer cells are involved in the cross-talk between cancer cells and other cells, and have an impact on the cancer progression and metastasis (Bose and Masellis, 2005; Kaminski et al., 2006). Furthermore, adipocytes were suggested to release factors that contribute to increase heart failure risk in overweight persons (Lamounier-Zepter et al., 2006) and have been shown to influence the steroidogenesis in vitro (Ehrhart-Bornstein et al., 2003). Conditioned media from a variety of cells derived from the central nervous system (CNS) affect p roliferation, survival and differentiation of neural stem/ progenitor cells (Taupin et al., 2000; Chang et al., 2003;Kaneko et al., 2003; Faijerson et al., 2006). In addition, an autocrine/ paracrine survival factor for cultured neural stem/progenitor cells has been found to stimulate neurogenesis in vivo (Taupin et al., 2000). These findings demonstrate that the identification of secreted proteins in conditioned media is important, since these factors can have potent effects on endogenous cells.
As opposed to traditional approaches that examine one or a few proteins at a time, proteomic strategies attempt to examine large numbers of proteins concurrently. Thus, one advantage with proteomic based analysis of conditioned media is that it allows for identification of proteins, which might not have been considered as interesting in a traditional hypothesis driven approach. Earlier proteomic studies on conditioned media have been performed on cells cultured, at least for some time, in serum-free media (Lafon-Cazal et al., 2003; Delcourt et al., 2005; An et al., 2006; Gronborg et al., 2006; Mbeunkui et al., 2006;Liu et al., 2008). The suppression of the lower abundance secreted proteins by the higher abundance serum proteins was thereby avoided. Furthermore, to differentiate the proteins identified in the serum-containing conditioned media as released proteins or proteins derived from the serum poses an analytical challenge. However, a drawback with this approach is that many mammalian cells require serum for optimal growth, and serum deprivation elicits several cellular responses. In many cell lines, apoptosis induced by serum deprivation is observed (Higuchi et al., 2006; Lebon et al., 2006; Wei et al., 2006). As a result, extra care has to be taken during culturing, since extracellular protein in the conditioned media due to autolysis is not desirable (Mbeunkui et al., 2006). Serum deprivation has also been demonstrated to affect the secretion profiles (Wei et al., 2006). Moreover, most data on cellular properties in cultures comes from culturing in serum-containing media making the results difficult to compare. As a consequence, proteomic strategies that facilitate identification of proteins released in serum-containing culture media have many advantages. In the present study a scratch injury model of reactive astrogliosis served as our model system. Lesioned astrocytes display many of the characteristic changes seen in reactive astrogliosis following an injury (Yu et al., 1993; Kornyei et al., 2000). Astrocytes belong to the glial cell group and are the most abundant cell type in the brain with many functions in central nervous system. Astrocytes are known to secrete numerous growth factors and extracellular matrix proteins modulating the activity of neighbouring cells and affecting cellto- cell interactions (Yoshida and Takeuchi, 1991; Schaar et al., 1993; Schwartz and Nishiyama , 1994; Aubert et al., 1995; Ridet et al., 1997; Faijerson et al., 2006). Many of the astrocytic properties are abnormally up-regulated in response to injury. Reactive astrogliosis is important for wound healing, but can also pose an obstacle to regeneration (Pekny and Nilsson, 2005). The role of reactive astrogliosis in healing or recovery in various central nervous system pathologies is still incompletely understood.
The aim of this study was to investigate whether metabolic labeling combined with mass spectrometric analysis could be used for identification of proteins, secreted by astrocytes, in serum-containing conditioned media. Stable isotope labeling by amino acids in cell culture (SILAC) has previously been used to identify differentially expressed proteins in both lysates from various cell systems (Ong et al., 2002; Everley et al., 2004; Ibarrola et al., 2004; Gruhler et al., 2005) and in serum- free conditioned media (An et al., 2006; Gronborg et al., 2006; Liu et al., 2008). In a SILAC experiment, cell cultures are grown in culture media containing either the normal amino acid or its isotopically labeled analogue. The amino acids substituted to the culture media are incorporated into the cellular proteins during growth. The two cell populations are mixed in equal ratios, and the quantitation of the proteins is based on the ion intensities of normal versus isotope labeled forms of the tryptic peptides, with identical sequences in the mass spectrometric analysis. In the present study, we take advantage of the fact that only the proteins originating from the cells contain the isotopically labeled amino acid. Accordingly, the released proteins can be distinguished from the medium proteins by their increased molecular mass corresponding to the labeled amino acid in the mass spectrometric analysis. The second objective of the study was to investigate whether the method allows for detection of differences in the secretion profiles between primary astrocytes and astrocytes in the scratch injury model of reactive astrogliosis. To our knowledge we are the first to identify secreted proteins in serumcontaining media using a proteomic approach involving SILAC and mass spectrometry.
Astroglial Cultures
Primary astroglial cultures were prepared from Sprague- Dawley rats (P1-2) as previously described (Nodin et al., 2005), with all experimental procedures approved by the Ethics Committee of the University of Göteborg. Briefly, rat pups were d ecapitated and the cerebral cortices were dissected and mechanically dissociated through 80-μm nylon meshes into Eagle’s minimum essential medium with 20% foetal calf serum (FCS, PAA Laboratories GmbH, Pasching, Austria), 2 mM L-glutamine and 1% penicillin/streptomycin (Life Technologies, Täby, Sweden). The medium had extra substances added to the following composition: 1.6 times the concentration of amino acids and 3.2 times the concentration of vitamins (Life Technologies), 48.5 mM NaHCO3 and 7.15 mM glucose (Merck, Darmstadt, Germany). After three days in culture, the medium was exchanged with a customised Eagle’s minimum essential medium deficient in all amino acids with 1.6 times the concentration of amino acids lacking arginine and 3.2 times the concentration of vitamins (Invitrogen) added. The medium also contained 20% dialysed foetal calf serum (FCS, PAA Laboratories GmbH, Pasching, Austria), 2 mM L-glutamine, 1% penicillin/streptomycin (Life Technologies), 48.5 mM NaHCO3 and 7.15 mM glucose (Merck). The cells were cultured in 5% CO2 at 37ºC in media containing either normal 12C6 14N4-arginine (= 98.5% purity, Sigma Aldrich, St. Louis, MO) or isotopically labeled 13C6 15N4-arginine (98% purity, Cambridge Isotope Laboratories, Andover, MA).
Confluent cultures (14-21 days after plating) were mechanically lesioned using a pipette tip in a 5 mm grid frame (Faijerson et al., 2006) and media were changed. Conditioned media were collected 48h after the injury was induced from both non-lesioned and lesioned astrocytes, filtered at 0.22 μm (Pall Corporation, East Hills, NY, USA) and immediately frozen at -80°C. After the conditioned media were collected, cells were harvested in ice-cold PBS supplemented with a protease inhibitor cocktail (Roche, Basel, Switzerland), 1 mM sodium orthovanadate (Sigma Aldrich) and 1 mM phenylmethanesulfonyl fluoride (Fluka, Sigma Aldrich). The samples were centrifuged (10 min, 4°C, 500 g), supernatants were discarded and pellets were stored at -80°C until usage. To validate the reactive astrogliosis model, cells were fixed (4% paraformaldehyde in PBS, 4°C, 10 min) 48h after the injury was induced and stained (1h, RT) with GFAP antiserum (rabbit, 1:500, DAKO, Glostrup, Denmark) in PBS containing 3% donkey serum (Jackson Immunoresearch Laboratories Inc., West Grove, PA) and 0.05% saponin (Sigma- Aldrich Sweden AB). Following three washes in PBS, cells were incubated for 1h at RT with the secondary antibody: Alexa Fluor 555-conjugated donkey anti-rabbit (1:2000, Molecular Probes) and the nuclear dye bisbenzimide from a stock at 5 ìg/ml (1:80, Hoechst 33258).
Protein Separation
Conditioned media (200 g) from lesioned astrocytes, and from lesioned and non-lesioned astrocytes pooled in equal ratios, were protein precipitated by adding ice-cold acetone, three times the volume of the conditioned media. The samples were mixed, stored at -20°C overnight, centrifuged (10 min, 4ºC, 35,000 g), and the supernatants were discarded. Lysis buffer (9 M Urea, 5% CHAPS and 0.05% SDS) supplemented with a protease inhibitor cocktail (Roche) was added to the cell pellets. The samples were sonicated (5x30 s), left in RT for 45 min and centrifuged (15 min, 4ºC, 35 000 g) before the supernatants were carefully removed. Protein concentrations were measured by the Bradford protein assay (BioRad, Hercules, CA, USA). Cell lysate (30 mg) from lesioned astrocytes, and from lesioned and non-lesioned astrocytes pooled in equal ratios, were treated with ReadyPrep (BioRad) according to the manufacturer’s instructions.
Protein pellets from conditioned media and cell lysates were dissolved in 25 mL of NuPAGE sample buffer (0.14 M Tris bas, 0.11 M Tris-HCl, 0.5 mM EDTA, pH 8.5, 10 % Glycerol, 75 mM LDS) and separation was performed on Nu- PAGE 10% Bis-Tris gels (Novex, San Diego, CA) and MOPS running buffer (50 mM MOPS, 50 mM tris, 3.5 mM SDS, 0.8 mM EDTA) at a constant voltage (200 V, 50 min). The gels were stained using Simply blue (Novex) to visualise the gel bands.
In-Gel Protein Digestion
The gel lanes of the conditioned media with an apparent molecular weight below 45 kDa were excised and divided into 12 equal sized sections. The whole gel lanes of the cell lysates were divided into 20 equal sized sections. Gel pieces were reduced and alkylated prior to digestion (Amanchy et al., 2005). The peptides were extracted with a 1:1 mixture of NH4HCO3 (50 mM) and acetonitrile (ACN), followed by a 1:1 mixture of 5 % formic acid and ACN twice. The resulting supernatants were pooled, dried and stored at -20ºC until analysis.
Mass Spectrometric Analysis
The peptide samples from the conditioned media and the cell lysate were resolved in 15 and 25 μl of aqueous 0.1% formic acid, respectively. The liquid chromatography mass spectrometry (LC-MS) and LC-tandem MS (LC-MS/MS) spectra were acquired using a hybrid Linear Ion Trap Fourier Transform Ion Cyclotron Resonance mass spectrometer equipped with a 7 T magnet (LTQ-FT, Thermo Electron Corp., Bremen, Germany) coupled to a Ettan™ MDLC (GE Healthcare, Uppsala, Sweden) multi-dimensional nanoflow chromatography system. The Ettan™ MDLC controlled by UNICORN software was run in the high-through put mode with two parallel flow paths for desalting and separation of p e ptides prior to on-line MS and MS/MS analyses. A Zorbax 300 SB-C18 trap column, (5 mm, i.d. 0.3 mm, 5 ì, Agilent Technologies, Palo Alto, CA) was used for online desalting and sample cleanup, followed by a nano-scale reverse phase column (Zorbax 300 SB-C18, 150 mm, i.d. 0.075 mm, 3.5 ì, ,Agilent Technologies) for high-resolution separation. After 17 minute linear run, loading the precolumn, the separation was performed at a flow rate of approximately 200 nL/min by applying a linear gradient of 0–60% B for 50 min. Mobile phase A consisted of HPLC-grade water with 0.1% formic acid while mobile phase B was 84% HPLC grade aqueous ACN with 0.1% formic acid. The injected volumes were 5 ìl. Each sample was subjected to two independent analyses using two separate, but identical analytical columns. The eluent was electrosprayed (+1.3 kV) from the emitter tip (i.d 10 ìm, uncoated, pre-cut, PicoTip™Emitter, New Objective, Inc., Woburn, MA, USA) into the heated capillary of the mass spectrometer. The mass spectrometer operated in the data dependent mode to switch automatically between MS and MS/MS acquisition. In the first experiment of the conditioned media both MS and MS/MS scans were performed within the linear ion trap (LTQ) to maximise the sensitivity of the analysis. Each scan cycle consisted of one full scan mass spectrum (m/z 300–1500) collected in profile mode and the three most abundant doubly and triply charged protonated ions in each MS-scan were selected for isolation, fragmentation and detection in the LTQ. The survey MS spectra (m/z 400– 1600) of the conditioned media sample were also acquired with the Fourier Transform Ion Cyclotron Resonance (FTICR) MS followed by MS/MS of the three most abundant doubly and triply charged ions in the LTQ. Dynamic exclusion was activated during 30 s with a repeat count of 1 in the analysis of the conditioned media. In the experiment of the cell lysate the survey MS spectra (m/z 300–1500) were acquired with the FT-ICR MS followed by MS/MS of the three most abundant doubly and triply charged ions in the LTQ. Dynamic exclusion was activated during 60 s with a repeat count of 1. The resolution was set to 100 000 for the FT-ICR.
Protein Identification and Quantitation
Acquired MS/MS data from the analysis of the conditioned media were submitted to database search using the inhouse Mascot software packet (Matrix Science, London, UK, (Perkins et al., 1999)). The data was searched against the NCBI database for protein identification. Database interrogation was; taxonomy (Mammals); enzyme (trypsin); variable modifications carbamidomethyl, oxidation of methionine residues and 13C6 15N4-Arg); mass values (monoisotopic); protein mass (unrestricted); peptide mass tolerance (1 Da); fragment mass tolerance (±0.4 Da), peptide charge state (2+ and 3+) and max missed cleavages (1). The peptide mass tolerance was set to 20 ppm in the search of data acquired with FT-ICR cell and rodent was selected in the search of the cell lysates. All spectra from one gel lane, run on the same analytical column, were searched as merged files, resulting in two independent data sets per sample. Proteins considered as identified proteins had at least two peptides with an individual mascot score corresponding to p<0.05 and p<0.1, respectively. The proteins identified in the conditioned media originated from Rattus Norvegius. Random search of the LCMS/ MS analysis of the conditioned media resulted in an average of 51 000 queries for each merged search, and only one false positive identified proteins with the given criterion. For proteins quantitation DeCyder MS Differential Analaysis software (DeCyderMS, GE Healthcare (Johansson et al., 2006; Thorsell et al., 2007)) was used. Acquired LC-MS raw data were converted and the PepDetect module was used for automated peptide detection, charge state assignments, and quantitation based on the peptide ions signal intensities in MS mode. The ratios of the integrated signal intensities were calculated for all the detected charged states of the unlabeled and labeled peptides ions in the SILAC pairs.
Protein Identification
The present study evaluated whether secreted proteins can be identified and differentiated from the medium proteins in serum-containing conditioned media by a combination of SILAC and mass spectrometry. Conditioned media from primary astrocytes and astrocytes in a scratch injury model of reactive astrogliosis substituted with either normal or isotopically labeled arginine served as our model system (Figure 1). The conditioned media samples were resolved by gel electrophoresis, revealing a distinct serum albumin protein band and a number of less intense protein bands (Figure 2). The gel lanes were excised and divided into equal sized sections, ingel digested followed by nano-scale LC prior to mass spectrometric analysis. This approach allowed for identification of proteins in the complex mixture of tryptic peptides and extensive separation of the proteins prior to proteolysis was not required, owing to the fact that peptides were isolated and fragmented individually in the mass spectrometric analysis.
Figure 1: Mechanically lesioned astrocytes displayed characteristics of reactive astrocytes. A-B: Confluent astrocytic cultures, derived from rat cortex, were mechanically lesioned. Following injury, bordering cells changed from a flat, polygonal shape (A), to a polarized morphology with processes extending towards the lesion (B). These polarized cells also displayed increased immunoreactivity for GFAP (red, dashed line indicates site of lesion). Cell nuclei were visualized using Hoechst 33258 (blue).
Figure 2: One-dimesional gels from the separation of the conditioned medium and the whole cell lysate. Serum albumin is the major component of the culture media with an apparent molecular weight between 45 and 65 kDa on the gel. The protein bands with an apparent molecular weight below the serum albumin band were selected for protein identification. The whole gel lane of from the cell lysate separation were further analysed for protein identification. There were no visible difference between lesioned astrocyte conditioned medium and the pooled conditioned medium, or for the cell lysate of lesioned astrocytes and pooled cell lysate (data not shown).
Most of the proteins identified in the conditioned media were bovine proteins derived from the serum. However, the database search also resulted in 22 proteins identified as proteins originating from rattus norvegicus in the serum-containing conditioned media (Table 1). For 12 of these proteins, at least one labeled arginine containing peptide was found, conf i rming that they were derived from the astrocytes rather than from the culture medium. The secreted proteins were differentiated from the medium components by the 10 Da mass differences of their fragment ions in the mass spectra corresponding to the incorporated arginine label (Figure 3). These results were confirmed with a second mass spectrometric analysis of the conditioned media (Table 1). However, the high mass accuracy obtained on the LTQ combined with FTICR was gained at the expense of loss in sensitivity compared to the analysis by the LTQ alone.
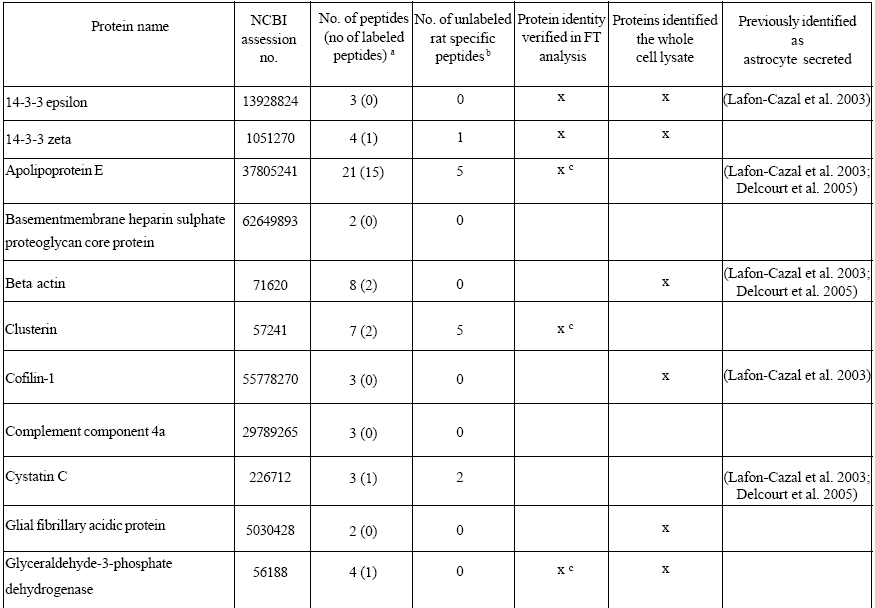
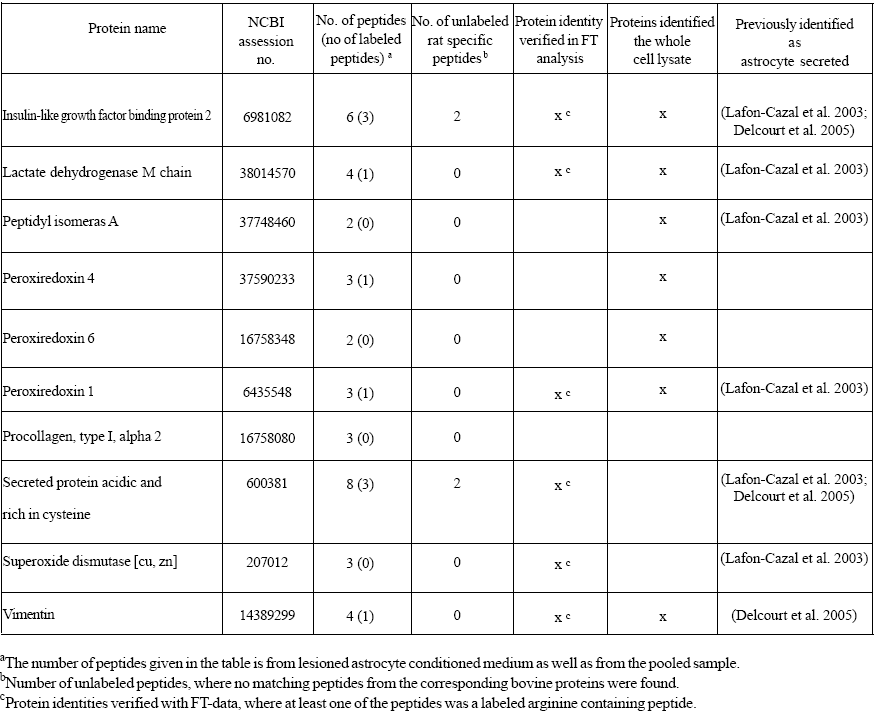
Table 1: Proteins identified in serum-containing astrocyte conditioned medium
Figure 3: Annotated fragmentation ion (CID) spectra for a SILAC pair in the conditioned medium from lesioned astrocytes. The protein was identified as glyceraldehyde-3-phospahte dehydrogenase. The peptides from the secreted protein were distinguished from the peptides from the corresponding proteins in the medium itself by the incorporated arginine label. The molecular masses of the precursor ions were 1555.8 and 1565.8 Da for the unlabeled and labeled peptides. The two fragmentation spectra are close to identical, except for the 10 Da mass differences owing to the labeled arginine. The amino acid sequence of the peptides was VPTPNVSVVDLTCR, obtained by interpretation of the fragmentation ions produced. The yseries of ions detected in the mass spectra are given in bold in the table.
The amino acid sequences of the proteins identified were compared with the sequences of the corresponding bovine proteins, in order to evaluate the possibility to discriminate astrocyte derived proteins from the serum proteins by their amino acid sequences. Only six of the identified proteins generated tryptic peptides detected in the mass spectrometric analysis with sequences different from the bovine proteins and a total of 17 unlabeled unique peptides were found for these proteins (Table 1). This is in contrast to the 32 peptides containing the arginine label found in the mass spectrometric analysis and the resulting 12 proteins that could be distinguished from the medium components. Thus, the incorporated arginine label was more effective in identifying secreted proteins in the serum-containing conditioned media than their amino acid sequences.
The released proteins in the conditioned media were also compared with the proteins identified in the whole cell extract. A number of proteins were identified in both the conditioned media and the cell lysate (Table 1). This finding strengthens the result that the proteins found in the conditioned media were derived from the astrocytes. Several of the proteins were only found in the conditioned media, suggesting that these proteins were enriched in the media. In the whole cell lysate, the level of these proteins was probably too low compared with the other relatively highly abundant proteins to allow for their identification.
The identified proteins in the serum-containing conditioned media included protease inhibitors (cystatin C), carrier proteins (insulin-like growth factor binding protein 2 (IGFBP- 2) and apolipoprotein E (ApoE)), antioxidants (Cu/Zn superoxide dismutase, peroxiredoxin 1, 4, 6), proteins involved in remodelling (clusterin, secreted protein acidic and rich in cysteine (SPARC)) and structural proteins (GFAP, vimentin, cofilin and beta-actin).
Protein Quantitation
A similar profile of released proteins was found in the conditioned media from lesioned and non-lesioned astrocytes. Neither were any significant quantitative differences between the two cell cultures found. However, this finding was a confirmation that the proteins in the lesioned astrocyte conditioned medium were secreted, rather than being the result of intracellular proteins leaking from ruptured cells. The quantitative analysis of a protein in the pooled conditioned media was based on the peptide ion intensities of the unlabeled and labeled peptides in the SILAC pairs. In (Figure 4) representative two-dimensional (2D) signal intensity maps of the LC-MS analysis used for relative quantitation and the LC-MS spectra of a peptide pair are shown. The 2D signal intensity map of the pooled sample illustrates that no quantitative differences could be found for the protein.
Figure 4: Signal intensity maps and ion chromatograms of the LC-MS analysis and LC-MS spectra of cystatin C peptides. (a) DeCyder MS displays LC-MS analyses as 2D signal intensity maps used for relative quantitation (Johansson et al., 2006). The insert shows m/z 880-895 and retention time between 22-40 min for lesioned astrocyte conditioned medium and the pooled sample. The intensity spots from the doubly charged SILAC peptide pair m/z 885.5 and 890.5 are indicated. The reactive astrocyte conditioned medium contains only the labeled peptide. Both the unlabeled and labeled peptides in the SILAC pair were found in the pooled sample. No significant difference in the concentration of cystatin C between the conditioned media was detected with DeCyderMS. The quantitative analysis was based on the normalised peptide ion signal intensities in the LC-MS separation. b) Extracted ion chromatograms for the cystatin C peptides in lesioned astrocyte conditioned medium (retention time 27.2 min) and in the pooled sample (retention time 27.4 min). The labeled peptide (m/z 890.5) was detected in the lesioned astrocyte conditioned medium. The chromatogram of the pooled sample shows that both the unlabeled (m/z 885.5) and labeled (m/z 890.5) peptides were found and that the peptides co-elute in the LC-separation.
Our results also revealed inherent limitations in the quantitative analysis of lesioned and non-lesioned astrocyte conditioned media. Surprisingly, the analysis of the lesioned astrocyte conditioned medium revealed both unlabeled and labeled peptides with the same amino acid sequence for some of the identified proteins (Figure 3). In the lesioned astrocyte conditioned medium only the labeled arginine containing peptide should be detected. The presence of both peptides could either be due to an inefficient labeling of the cellular proteins or that the unlabeled tryptic peptides originate from the medium components. In order to determine the labeling efficiency, the whole cell lysate of the lesioned astrocytes was analysed. The analysis showed close to complete incorporation of the labeled amino acid into the proteins, since peptides containing the unlabeled amino acid were nearly absent in the MS-spectra. This finding confirmed that the unlabeled proteins were present in the culture medium itself and that several of the secreted proteins generated tryptic peptides with the same amino acid sequences as the corresponding bovine proteins. Consequently, the signal ion intensity from the unlabeled peptides in the pooled sample included both proteins released from the astrocytes and from the culture medium itself, which complicated the quantitative analysis for these proteins. To circumvent this drawback and to calculate a correction factor to obtain the correct expression ratios, the intensity ratios of both lesioned astrocyte conditioned medium and the pooled samples were calculated. An estimation of the contribution of the medium components to the unlabeled peptides in the SILAC pair was thereby obtained. However, despite these analyses no quantitative differences were observed for these proteins. Furthermore, co-elution of different peptides with close to the same m/z during the LC-run also restricted the quantitative analysis. Several of the SILAC pairs identified in the fragmentation analysis that could not be quantified had interfering peptides with similar physicochemical properties that eluted closely enough to overlap in the LC-separation (Figure 5). Moreover, the quantitative analysis was limited by the generally low number of detected SILAC peptide pairs. To be able to calculate statistically significant intensity ratios, it is essential to obtain multiple SILAC pairs from each protein.
Figure 5: Signal intensity map and ion chromatograms of a SILAC peptide pair identified as the 14-3-3 zeta protein in the lesioned astrocyte conditioned medium. a) The insert shows m/z 770-785 and retention time between 12-30 min for lesioned astrocyte conditioned medium. The intensity spots from the doubly charged SILAC peptide pair m/z 774.4 and 779.4 are indicated on the map. Both the peptides in the SILAC pair were identified in the fragmentation analysis of the lesioned conditioned medium. The unlabeled peptide in the pair is present in the medium itself and is not derived from the cells. The labeled peptide has an interfering peptide with close to the same m/z that eluted at almost the same time. b) DeCyder MS ion chromatograms for the unlabeled and labeled peptides in the SILAC pair. Only the unlabeled peptide in the SILAC pair was detected and identified with DeCyderMS. Both identification of the labeled peptide and calculation of the ion intensity ratio in the quantitative analysis were prevented by co-elution of the interfering peptide and the labeled peptide. The ion chromatogram of the labeled SILAC peptide shows two superimposed elution curves, illustrating the overlap of the two peptides in the LC-MS separation. The mass difference between the two peptides co-eluting is approximately 3 mass units, and their isotopic envelopes are not resolved from each other in the LTQ-MS spectrum.
Metabolic labeling and mass spectrometry allow for identification of cell derived proteins in serum-containing conditioned media. The employed strategy was different from previous studies of astrocyte secreted proteins, where the cells were cultured under serum-free conditions during the secretion period. The proteomic strategy utilized in these studies was 2D gel electrophoresis followed by peptide mass fingerprinting of selected protein spots for identification (Lafon-Cazal et al. , 2003; Delcourt et al., 2005). However, many mammalian cells require serum for optimal growth, and serum deprivation elicits several cellular responses. Moreover, most data on cellular properties comes from culturing in serum-containing media making the results difficult to compare. Subsequently, analytical methods that facilitate the study of secreted proteins in serum-containing conditioned media are valuable and have many advantages. The fact that the presence of serum in the conditioned media increases the risk of proteins co-migrating into the same spot on the 2D gel makes traditional 2D gel electrophoresis unsuitable for analysis of serum-containing media. On the other hand, metabolic labeling and mass spectrometric analysis do not require extensive separation prior to proteolysis, since the proteins can be identified and quantified in complex tryptic digest. Additionally, proteins derived from cells can also easily be differentiated from the medium proteins in the mass spectrometric analysis by the incorporated label.
As expected most of the identified proteins in the conditioned media were bovine proteins. However, the database search also resulted in a number of proteins identified as proteins originating from rattus norvegicus and for 12 of these proteins at least one labeled peptide was found. The peptide amino acid sequences of the proteins identified were also compared with the sequences of the corresponding bovine proteins. Notably, the comparison revealed that the incorporated arginine label was more efficient in discriminating secreted proteins from the corresponding bovine proteins in the conditioned medium than their amino acid sequences.
Our results lend support to previous studies of culture media conditioned by primary astrocytes (Lafon-Cazal et al., 2003; Delcourt et al., 2005). A number of the proteins were identified as released by astrocytes under serum-free conditions during the secretion period with proteomic strategies involving mass spectrometry (Lafon-Cazal et al., 2003; Delcourt et al., 2005) and strengthen our results that the proteins were indeed secreted. However, we also identified a number of proteins, including clusterin, not previously found in these earlier studies. Clusterin was identified in conditioned media from both lesioned and non-lesioned astrocytes. This glycoprotein is highly expressed in response to tissue injury and plays a central role in CSN remodeling after ischemic damage (Imhof et al., 2006). Clusterin also displays neuroprotective properties and the increased expression of the protein can limit the damage following ischemia (Wiggins et al., 2003).
A similar profile of secreted proteins and no significant quantitative differences were found in the conditioned media from the two cell cultures. One explanation for the similarities in qualitative and quantitative secretion profiles could be that the proteins identified were the major proteins released by astrocytes, and do not differ significantly between the two conditions. However, this finding was a confirmation that the proteins in the lesioned astrocyte conditioned medium were secreted, rather than being the result of intracellular proteins leaking from ruptured cells. Proteins following lysis in the conditioned media were limited by changing of the culture media after induction of injury, and the conditioned media for proteomic analysis were collected after only 48h of further culturing. Moreover, it is likely that some differences in protein expression were masked by the fact that only a fraction of the cells in the astrogliosis model was lesioned and therefore directly affected. It should also be noted that non-lesioned primary astrocytes, derived from neonates, have been suggested to display a reactive phenotype since they recapitulate many of the expression patterns seen in reactive astrocytes in the injured adult brain (Wu and Schwartz , 1998). Thus, a different model system might have been more suitable in the evaluation whether the combination of SILAC and mass spectrometry can detect quantitative differences in the secretion profiles in serum-containing conditioned media.
The present approach can be optimised in various ways to enhance its ability to identify and quantify released proteins in serum-containing conditioned media. The number of labeled peptides found in the mass spectrometric analysis is crucial for both identification and quantitation of the secreted proteins. In this study, only the tryptic peptides containing arginine were labeled and as a consequence less then half of the total number of tryptic peptides were theoretically labeled. Consequently, the capability of this method to identify and quantify secreted proteins by the incorporated label would be significantly improved by using both isotopically labeled arginine and lysine and thereby maximising the number of tryptic peptides.
A reduction in the sample complexity prior to LC-MS analysis also has this effect, since the number of peptides eluting within the same time window is reduced. Peptide overlap in the 2D signal intensity maps causes both errors in measurements of the intensity ratios and suppression of relatively low abundance proteins by more abundant ones. One advantage with SILAC experiments is that the use of any method of protein purification to reduce the complexity is allowed, without introducing errors into the final quantitative analysis. Albumin removal might be an attractive strategy to reduce sample complexity and to enrich the secreted proteins compared to the original sample. As a consequence of the albumin removal, the quantity of secreted proteins in gel electrophoretic separation can be significantly increased. In the present study the protein bands below the serum albumin protein band were excised and in-gel digested for analysis. Consequently, only a part of the astrocytic secretome was studied. Despite this limitation the database search resulted in 22 proteins originating from rattus norvegicus and 12 of these were confirmed as derived from the cells by at leased one labeled peptide. The number of identified proteins would probably have be significantly increased by an approach involving albumin removal. However our result demonstrates that secreted proteins can be identified in serum-containing conditioned media. Another strategy to reduce the sample complexity can be fractionation followed by LC-MS analysis. We have previously shown that prefractionation using micro-scale isoelectric focusing increased the number of proteins identified drastically and improved their quantitation (Thorsell et al., 2007). This is a result of the combination of increased protein load, the enrichment of proteins as well as the reduced sample complexity relative the unfractionated sample. Moreover, the quantitative limitation due to the secreted proteins generating tryptic peptides identical with the corresponding bovine proteins can be circumvented by labeling both the cell cultures with different isotopically labeled amino acids yielding different mass shifts (e.g. 13C6 14N4-Arg and 13C6 15N4-Arg). This would make it possible to distinguish the extracellular proteins from both the cell cultures from the medium components.
Our results demonstrate that SILAC in combination with mass spectrometric analysis allows for the identification of the secreted proteins in serum-containing conditioned media. The released proteins were distinguished from the medium components by the labeled amino acid incorporated into the cellular proteins during culturing. This approach can be used as an initial screening tool to identify proteins released in serum-containing conditioned media. Importantly, it can be applied to all cultured cells where it is desired to study their secretome.
The authors like to gratefully acknowledge support of this project by The Sahlgrenska University Hospital, Göteborg, Sweden, The Inga-Britt and Arne Lundberg Research Foundation, Göteborg, Sweden, Swedish Medical Research Concil (12575, 12335) and Aina Wallströms och Mary-Ann Sjöbloms stiftelse för medicinsk forskning, Göteborg, Sweden.