Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2019)Volume 12, Issue 8
A growing body of evidence has highlighted that a wide range of plant processes including growth, photosynthesis, and stress response are regulated by 595 nm light. However, the molecular mechanisms underlying 595 nm-induced signals are not known. The aim of this work was to study the comparative proteomic changes in leaves of Arabidopsis thaliana Col-0 plants treated with narrow-wavelength 595 nm light or fluorescent light for 5 days. Harvested plant samples were analyzed using inline RP-SCX-RP liquid chromatography coupled with LTQ mass spectrometer, resulting in the identification of 1538 proteins. Linear regression modeling of proteins’ relative abundance revealed a total of 23 differentially abundant proteins (DAPs). Functional analysis of these DAPs demonstrated the role of several biological mechanisms in A. thaliana’s response to 595 nm light including stress response and metabolic processes. A network analysis of these DAPs revealed the importance of energy and redox regulation mechanisms. Further analyses determined potentially important roles for proteins associated with glycolysis, ATP synthase complex, cell wall modification, and thylakoid membrane that may modulate the plant’s adaptive response to 595 nm light. A significant enrichment of DAPs for PSII tolerance capacity, as well as associated Ca2+ and ROS signaling pathways were also identified. Collectively, this study provides an important insight into potential molecular pathways that sustain a plant’s response to 595 nm light.
Adaptation; Arabidopsis thaliana; Light Wavelength; Protein Network; Quantitative Proteomic; Stress Response; Tolerance Response; Mass Spectrometry; Liquid Chromatography; Light Emitting Diode LED
The spectral quality of the radiation, adopted by plant production systems, efficiently drives plant growth and development [1]. It is therefore possible to influence plant adaptation and tolerance by controlling light-signal mechanisms to significantly improve agricultural production [2]. For example, a five-fold increase in annual greenhouse cucumber productivity from 50 to 250 kg/ m2 was reported by controlling the spectral light climate. With the increasing effects of climate change and a rapidly increasing world population, controlled environment agriculture is a promising solution to these global challenges [3]. Understanding the mechanisms that underlie plant spectral light responses and tolerance is hence paramount to improve plant agriculture productivity and global food security. Within the photosynthetically active radiation (PAR) light spectrum (400–700 nm), plant photoreceptors perceive the major wavelengths corresponding to blue (400–500 nm) and red (600–700 nm) regions, and less within the green (500–600 nm) [4]. While only 10–50% of these regions are reflected by plant chloroplasts [5], there is a misconception that plants do not make use of the wavelengths regions (500–600 nm) [6].Within this region, there is a narrow wavelength with a peak at 595 nm that a growing body of evidence has highlighted for its physiological impact on plants growth and photosynthesis [7,8]. For example, reported consequences of 595 nm include reduction in growth, chlorophyll content, photochemical quenching and quantum efficiency of PSII photochemistry. This wavelength is weakly absorbed by photosynthetic pigments [5], and 595 nm wavelength is arguably a less efficient light energy source for driving photosynthesis [9-12]. Congruently, under 595 nm, the activity of Rubisco decreases which may be due to a reduction in photosynthesis. Plant illumination with 595 nm results in elongation and less leaf area. Furthermore, a decrease in CO2 fixation and net photosynthesis, leading to reduced growth and yield has been reported [11,12]. However, McCree showed that 595 nm light exhibits some of the highest photosynthetic activity of any of the other wavelengths of light tested [13]. Additionally, 595 nm light plays a highly regulatory role in abundance and activity of key proteins that mediate stress tolerance in plant [14]. For instance, 595 nm light induces the abundance/activity of antioxidant enzymes such as superoxide-dismutase (SOD), catalase (CAT), and peroxidase (POD), ultimately reducing the accumulation of free radicals. Thus, 595 nm light regulates an integrated molecular network at multiple levels to bring about tolerance responses. Despite being a probable light source for understanding regulatory signals in plant growth and development related to stress and tolerance responses, the molecular mechanisms and impact on plant growth underlying the effect of 595 nm light remain unclear. Plants have developed a sophisticated sensory system to optimally grow and regulate numerous developmental processes under ambient light quality conditions [15,16]. A variety of receptor molecules, often proteins, are involved in monitoring radiation, and consequently transduce signals to trigger a response [17]. The signal-response network often involves complex interactions between signal transduction and many different processes to regulate translational responses and protein expression throughout the whole plant [18]. Proteomic techniques are a powerful means of detecting quantitative variations in relative protein abundance and fundamental cellular biological processes in response to a defined set of environmental conditions [19]. To investigate the molecular mechanism underlying plant responses to 595 nm light, a RP-SCXRP liquid chromatography coupled with LTQ mass spectrometer was applied. As studies have demonstrated that plant growth, photosynthesis and developmental responses to 595 nm light are directly opposed to the normal progression of light-mediated processes, we compared the changes in proteins abundance of the model plant Arabidopsis thaliana treated under 595 nm light and fluorescent light (FL),as the control. The obtained data indicate that many proteins are differentially expressed (activated or repressed) in plants grown under 595 nm light. Enriched proteins were involved in photosynthesis, metabolic processes, stress response, and ROS signaling. This study highlights light wavelength-regulated molecular networks and potential pathway crosstalk in plants tolerance response under 595 nm light.
Plant material and growth conditions
Seeds of the Arabidopsis thaliana accession Col-0 were obtained from the Arabidopsis Biological Resource Center (ABRC; https://www.arabidopsis.org). Seeds were dark-incubated at 4°C. After 2 days, germinated seeds were placed in rockwool cubes (Grodan A/S, DK-2640, Hedehusene, Denmark) and grown hydroponically for 21 days at a photosynthetic photon flux density (PPFD) of 70 μmol·m-2·sec-1 of fluorescent lamps (FL; 4200 K, F72T8CW, Osram Sylvania, MA, US) (Figure 1A) under a 16 h photoperiod in a growth chamber (TC30, Conviron, Winnipeg, Canada). The low light PPFD (70 μmol·m-2·sec-1) was used to limit light-induced stress responses [20]. Seeds density was adjusted to prevent 21-day old plant leaves from shadowing each other. PPFD was measured over the growing area (49 cm × 95 cm) using a grid size of 3 cm2 for each reading to provide uniform distribution of light over the growing surface. Fresh half-strength Hoagland nutrient solution [21] was provided every other day.
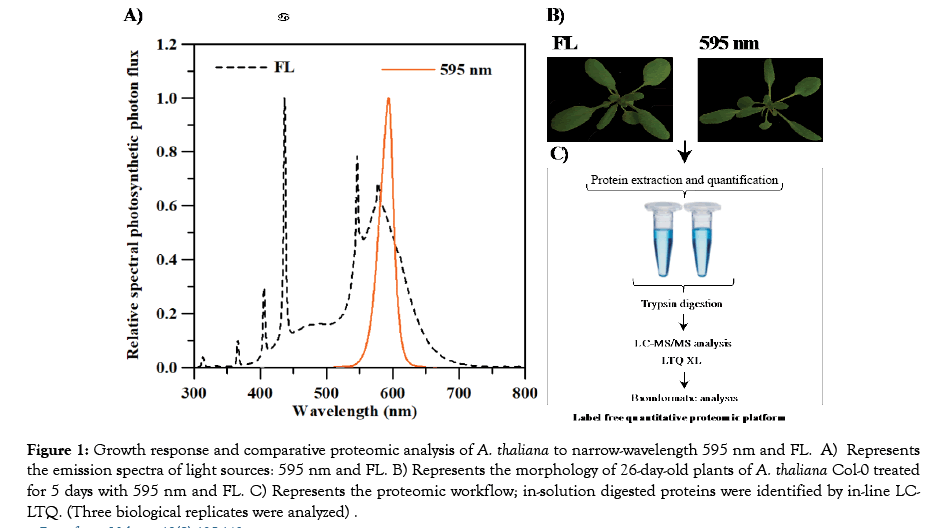
Figure 1: Growth response and comparative proteomic analysis of A. thaliana to narrow-wavelength 595 nm and FL. A) Represents the emission spectra of light sources: 595 nm and FL. B) Represents the morphology of 26-day-old plants of A. thaliana Col-0 treated for 5 days with 595 nm and FL. C) Represents the proteomic workflow; in-solution digested proteins were identified by in-line LCLTQ. (Three biological replicates were analyzed) .
Light treatments
A customized light emitting diode (LED) array with a precisely measured single-emission narrow peak of 595 nm (VanqLED, Shenzhen, China) was used for light treatments (Figure 1A). After 21-day growth under FL, 80 plants were randomly divided into two groups and treated for 5 days under 595 nm LED or FL light of a wide spectral emission range (400–700 nm) with the following environmental conditions: 16 h photoperiod, 23/21°C (day/night), 50% relative humidity, and ambient CO2 levels. Wavelength spectra of the LED and FL lights were measured using a PS-300 spectroradiometer (Apogee, Logan, UT, US). After 5 days of treatment, 30 plants were randomly harvested as a single sample, immediately frozen in liquid nitrogen, and stored at -80°C for protein extraction. The above-mentioned experiments were repeated three times to provide three biological replicates.
Protein extraction and digestion
Frozen samples were ground in liquid nitrogen using a mortar and pestle, prior to protein extraction and digestion [22], adapted here for A. thaliana. Briefly, 100 mg of ground plant tissue was homogenized on ice in 100 μL lysis buffer (Tris/10 mM CaCl2 at pH 7.6 and 6 M guanidine/10 mM DTT) (Sigma-Aldrich Canada, Oakville, ON). The solution was boiled for 5 min and vortexed every 2 min for the first hour before being incubation at 37°C for 12 h. Solubilized mixtures were sonicated in an ice bath with 40% amplitude for two cycles of 60 s each, with a 30 s cool-down period between cycles (Crystal Electronics, Stamina-XP Ultrasonicator, Newmarket, ON, Canada). The protein extract was boiled for 5 min prior to centrifugation (3000 g) at 25°C for 15 min (Beckman Avanti J25-I, Brea, CA). Finally, the protein concentrations in the lysates were quantified using a BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA, US). Aliquots containing 200 μg protein were reduced with 25 mM DTT, followed by protein precipitation with trichloroacetic acid. Samples were centrifuged at 4500 g for 10 min before washing with ice-cold acetone to remove lipids and excess SDS. Pellets were re-suspended in 250 ul of 8 M urea, 100 mM Tris-HCl, pH 8.0 using sonic disruption and incubated for 30 min at room temperature. Denatured proteins were reduced with 5 mM DTT. To prevent reformation of disulfide linkages, cysteines were blocked with 20 mM iodoacetamide.
Sequencing-grade trypsin (Promega, Madison, WI, US) was diluted with 50 mM Tris/10 mM CaCl2. Cysteine-blocked protein pellets were suspended in two aliquots of sequencing-grade trypsin (1:50 w/w), then incubated overnight at 37°C, followed by the addition of a second aliquot of trypsin and incubation for 4 h, followed by the addition of 1 M DTT to a final concentration of 20 mM. Following digestion, peptide samples were centrifuged for 10 min at 4500 g (VWR Costar model V, Radnor, PA) and pellets were discarded. The supernatant was adjusted to 200 mM NaCl, 0.1% formic acid and filtered through an Ultrafree- MC 45 μm spin filter (Ultrafree-MC UFC30HV00, pore size 0.45 μm, EMD Millipore, Billerica, MA). The peptide concentration was quantified using a BCA Protein Assay Kit before being stored at -80°C until mass spectroscopy (MS) analysis.
LC-MS/MS
Peptide analysis was conducted using inline three phasic (RP-SCXRP) HPLC coupled to a linear ion trap mass spectrometer (LTQ, Thermo Fisher Scientific, San Jose, CA, USA) [23]. A total of 50 μg peptides were loaded onto an in-house, packed 75 μm inner diameter biphasic back column containing ~3–5 cm strong cation exchange resin for charge-based separation of peptides followed by ~3–5 cm C18 RP for online washing and removing residual urea and NaCl (Luna 5 μm 100A and Aqua 5 μm 100A, Phenomenex, Torrance, CA) [24]. Each loaded column was first washed for 30 min off-line with LC–MS grade H2O (O.1% F.A) to remove salts before being placed in-line with a front column including a nanospray emitter tip (150 μm with 15 μm tip; New Objective, Woburn, MA). The front column was packed with ~15–20 cm of C18 RP resin for hydrophobicity-based separation of peptides, adapted from previously described methods [24]. The analysis was conducted via LC-MS/MS in 12 steps of a 5–30% ACN gradient and 0.125% FA over 180 min at a flow rate of 400 nl/min. Data were acquired in a positive mode by Data-Dependent Acquisition (DDA) operated by the software Xcalibur (v2.0.7) (Thermo Fisher Scientific, CA, USA). Survey scan was followed by the CID MS/ MS and high energy collision dissociation MS/MS of the 5 most intense ions in the LTQ for CID at 35 normalized collision energy, 3 m/z isolation width, 10 ms activation time, and previously fragmented ions were dynamically excluded for 30 s.
Protein identification
For each light condition, three biological replicates were tested. By injecting the same sample either one or two times as technical replicates; as a result, six and five samples were generated overall for the 595 nm light and FL control samples, respectively. The acquired LC-MS/MS spectra were extracted from Thermo RAW files that corresponded to each biological or technical replicate. Raw files were converted into MS2 files using RawXtract software, followed by conversion into mass lists using the Raw Extract 1_9_8 program [25]. The raw data files were processed and quantified using PatternLab for Proteomics software (v4.0.0.62) (available at: http://www.patternlabforproteomics.org/) [26]. Peptide sequence matching (PSM) was performed using the Comet algorithm [27] against the UniProt database (http://www.uniprot.org/) with A. thaliana protein entries downloaded August 2017, containing mitochondria, chloroplast proteins, and common contaminant proteins (i.e. bovine trypsin and human keratin). The search considered semi-tryptic peptide candidates. The oxidation of methionine was considered as a variable modification. The Comet search engine considered a precursor mass tolerance of 450 ppm. Because of the low resolution of LTQ-XL technology, we performed a second round of search to ensure the robustness of the result. To this aim, MS2 files were searched with ProLuCID search engine [28] against a sequence database used for PatternLab’s Comet search engine. Parameters used for performing the searches were semi-tryptic hydrolysis allowing up to two missed cleavages, for both precursor tolerance and fragment tolerance of 500 ppm. The PSM provided by ProLuCID were subsequently filtered through the PatternLab's Search Engine Processor (SEPro) module [29]. The same conclusion was reached using these two search engines. The default on SEPro for both data was set on low resolution MS1 and experiment with more than 50k spectra. All identification results are reported with <1% FDR at the peptide level based on the number of labeled decoys. Spectral counts were normalized using normalized spectral abundance factor (NSAF) approach that is widely used for the quantitative analysis of label free proteomics experiments [30]. Normalized spectral counts for each protein was next compared between the two light conditions using the T-Fold approach implemented in PatternLab software [31]. Proteins were deemed as differential abundant by setting two criteria: minimum fold change of 1.5, a Benjamini-Hochberg corrected p-value (FDR) of 0.05. To further confirm the significance of identified differentially abundant proteins, we compared their fold changes of the differentially abundant proteins under 595 nm in an independent dataset [32]. To assess the significance of the results, iterating 10000 times, we randomly permutated the fold change patterns in the independent dataset. Empirical p-value was next estimated by comparing the number of differentially abundant proteins that are consistently up or down regulated in both datasets with the distribution of what that is expected by chance.
Function annotation and classification of the DAPs
Protein-protein interaction (PPI) networks were performed for all differentially abundant proteins using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. The data setting for PPI construction was set as follows; organism: Arabidopsis thaliana, and minimum required interaction score: highest confidence (0.7). The hub node identification was conducted using Cytoscape 3.4.0 [33]. Enrichment analysis for biological process annotations were performed on DAPs using the BiNGO database databank available as a tool inside Cytoscape [33]. Enrichment analyses for metabolic pathway annotations were conducted on DAPs using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database as a tool inside STRING.
A. thaliana Col-0 growth response to narrow-wavelength 595 nm light
Twenty-one-day-old A. thaliana Col-0 grown from seed under FL light were treated with 595 nm LED light or FL (Figure 1A) for 5 days. After treatments, plants treated with 595 nm LED light showed morphological changes indicative of weak light absorption, including smaller leaves with longer petioles, when compared with control plants treated with FL (Figure 1B).
A. thaliana Col-0 proteomic response to narrowwavelength 595 nm light
To investigate proteomic changes in plants response associated with 595 nm light, LC-MS/MS analysis was performed on proteins extracted from leaves of A. thaliana Col-0 plants treated with 595 nm or FL light for 5 days (Figure 1C). LTQ certified the presence of 991 proteins, detected with at least two unique peptides across all samples. Of these identified proteins, a total of 334 proteins were detected three times through proteomic analysis of three independent biological replicates for each light condition. Proteins with significant abundance changes (fold change ≥ 1.5 and FDR ≤ 0.05) in response to 595 nm versus FL are listed in Table 1. A total of 23 unique proteins met the criteria that indicated differentially abundant proteins (DAPs). Among these DAPs, expression of 14 proteins was up-up-regulated, while the expression of 9 other proteins was down-regulated, when treated with 595 nm light.
| No.a | Protein numberb | Protein namec | Protein Description | Sub-localizationd | Fold change | p-value |
|---|---|---|---|---|---|---|
| Upe | ||||||
| Photosynthesisf | ||||||
| 1 | P56778 | PSBC | PSII CP43 reaction center | CY | 1.81 | 0.041 |
| 2 | Q01908 | ATPC1 | ATP synthase gamma chain 1 | CL | 1.57 | 0.029 |
| Carbon and amino acid metabolism | ||||||
| 3 | Q42472 | GAD2 | Glutamate decarboxylase 2 | CY | 6.83 | 0.012 |
| 4 | P10795 | RBCS-1A | Ribulose bisphosphate carboxylase small chain 1A | CL | 2.36 | 0.033 |
| 5 | O50008 | MS1 | 5-methyltetra-hydropteroyl triglutamate-homocysteine methyltransferase 1 | CY | 2.16 | 0.023 |
| 6 | P48491 | TPI | Triosephosphate isomerase | CY | 1.93 | 0.031 |
| 7 | Q9SRV5 | MS2 | 5-methyltetra-hydropteroyl triglutamate-homocysteine methyltransferase 2 | CY | 1.8 | 0.01 |
| 8 | P25696 | ENO2 | Bifunctional enolase 2/ transcriptional activator | N | 1.62 | 0.023 |
| Cytoskeleton and cell wall | ||||||
| 9 | O49006 | PME3 | Pectinesterase/pectinesterase inhibitor 3 | CW | 2.48 | 0.008 |
| Protein synthesis, folding and degradation | ||||||
| 10 | P22953 | HSC70-1 | Probable mediator of RNA polymerase II transcription subunit 37e | N | 5.61 | 0.034 |
| 11 | Q94K05 | CCT8 | T-complex protein 1 subunit theta | CY | 3.63 | 0.043 |
| Transportation | ||||||
| 12 | Q9SJT9 | AT2G21390 | Coatomer subunit alpha-2 | CY | 2.54 | 0.047 |
| 13 | P31167 | AAC1 | ADP, ATP carrier protein 1 | M | 2.04 | 0.006 |
| Signaling and defense | ||||||
| 14 | Q9SYT0 | ANN1 | Annexin D1 | PM | 2.98 | 0.03 |
| Down | ||||||
| Photosynthesis | ||||||
| 15 | Q94BS2 | MET1 | Protein MET1 | CL | 0.39 | 0.036 |
| 16 | Q9C9I7 | PSB33 | Rieske (2Fe-2S) domain-containing protein | CL | 0.32 | 0.033 |
| Carbon and amino acid metabolism | ||||||
| 17 | Q39161 | NIR1 | Ferredoxin-nitrite reductase | CL | 0.5 | 0.038 |
| 18 | Q9LF98 | FBA8 | Fructose-bisphosphate aldolase 8 | CY | 0.34 | 2E-04 |
| Protein synthesis, folding and degradation | ||||||
| 19 | Q9SZD6 | EF-Ts | Elongation factor Ts | M | 0.46 | 0.014 |
| Transportation | ||||||
| 20 | Q9FMF7 | DIT2-1 | Dicarboxylate transporter 2.1 | CL | 0.58 | 0.044 |
| Signaling and defense | ||||||
| 21 | Q9ZUC1 | AOR | Quinone oxidoreductase-like protein At1g23740 | CL | 0.55 | 0.043 |
| 22 | Q9C5R8 | AT5G06290 | 2-Cys peroxiredoxin BAS1-like | CL | 0.35 | 0.03 |
| 23 | Q949U7 | PRXIIE | Peroxiredoxin-2E | CL | 0.28 | 0.043 |
aNumerical list of 595 nm responsive proteins; bProtein number and the abbreviation commonly used for the protein. cProtein name given by Uniprot_Arabidopsis database. dSubcellular localization of each protein given by the PPDB database. eProtein expression pattern; upregulated and downregulated. fFunctional group. CL: Chloroplast; CW: Cell Wall; CY: Cytoplasm; N: Nucleus; PM: Plasma Membrane; PSII: Photosystem II.
Table 1: List of differentially abundant proteins in A. thaliana treated with narrow-wavelength 595 nm.
Functional annotation of differentially abundant proteins in A. thaliana Col-0 treated with narrow-wavelength 595 nm light
To identify mechanisms involved in the plant response to narrowwavelength 595 nm light, we annotated the DAPs via the GO function and KEGG pathway enrichment analysis using BiNGO and the STRING database, respectively. A total of 143 GO terms and 10 KEGG terms were identified with Benjamini-Hochberg corrected false discovery rate (FDR) <0.05 (Figure 2; Table 2). The enriched GO terms were associated with various biological processes, including response to stimulus, response to stress, and small molecule metabolic processes. The overrepresented GO cellular component terms included chloroplast, membrane, and cytoplasm. The enriched GO molecular function categories were ion-binding and catalytic activity. The major KEGG pathways included metabolic pathways (10 DAPs), biosynthesis of secondary metabolites (6 DAPs), and biosynthesis of amino acids (5 DAPs). The GO and KEGG analyses provide an overarching view on the associated biological processes involved in plant response to narrow-wavelength 595 nm light.
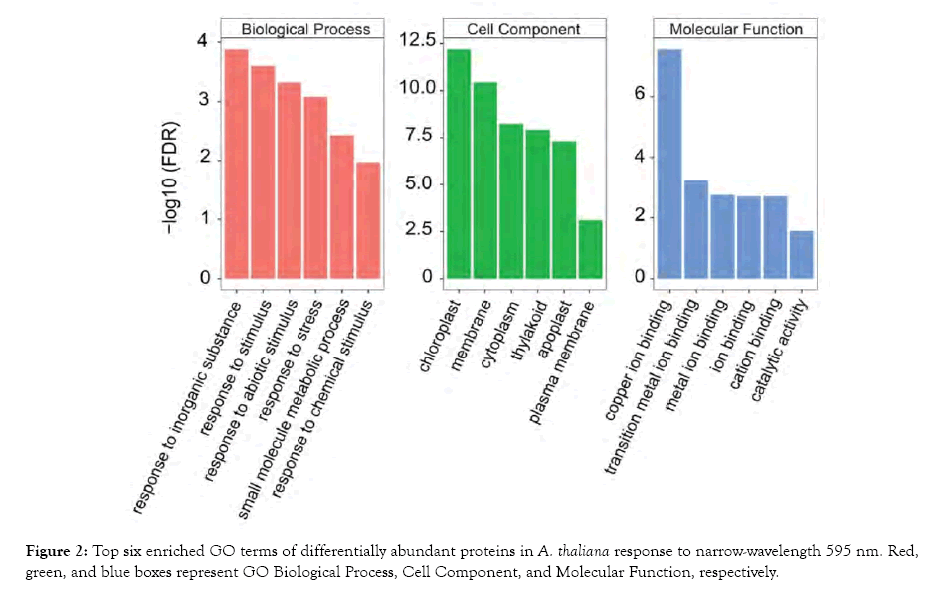
Figure 2: Top six enriched GO terms of differentially abundant proteins in A. thaliana response to narrow-wavelength 595 nm. Red, green, and blue boxes represent GO Biological Process, Cell Component, and Molecular Function, respectively.
| #term IDa | Term description | Countb | FDR | Matching proteins |
|---|---|---|---|---|
| ath01100 | Metabolic pathways | 10 | 4.94E-05 | MS1, ATPC1, MS2, FBA8, GAD2, ENO2, PME3, PSBC, RBCS-1A, TPI |
| ath01230 | Biosynthesis of amino acids | 5 | 4.94E-05 | MS1, MS2, FBA8, ENO2, TPI |
| ath00710 | Carbon fixation in photosynthetic organism | 3 | 0.00027 | FBA8, RBCS-1A, TPI |
| ath01200 | Carbon metabolism | 4 | 0.00047 | FBA8, ENO2, RBCS-1A, TPI |
| ath00010 | Glycolysis/gluconeogenesis | 3 | 0.00066 | FBA8, ENO2, TPI |
| ath00450 | Selenocompound metabolism | 2 | 0.00066 | MS1, MS2 |
| ath01110 | Biosynthesis of secondary metabolites | 6 | 0.00087 | MS1, MS2, FBA8, GAD2, ENO2, TPI |
| ath00051 | Fructose and mannose metabolism | 2 | 0.0046 | FBA8, TPI |
| ath00195 | Photosynthesis | 2 | 0.0056 | ATPC1, PSBC |
| ath00270 | Cysteine and methionine metabolism | 2 | 0.011 | MS1, MS2 |
aPathway ID given by KEGG database. bNumber of the involved proteins in protein set. FDR: False Discovery Rate.
Table 2: KEGG pathways enriched by differentially abundant proteins in A. thaliana response to narrow-wavelength 595 nm.
Molecular networks involved in A. thaliana Col-0 response to narrow-wavelength 595 nm light
To discern the interaction networks for the DAPs identified in A. thaliana response to narrow-wavelength 595 nm light, a proteinprotein interaction network (PPI) was constructed using the STRING database (Figure 3). This analysis demonstrated that the identified DAPs are highly interconnected. Core central proteins, with high connections in the PPI network, were analyzed using the Cytoscape database. Hub analysis of the network suggested ATPC1, FBA8, and AT5G06290 were central proteins in these networks and in the plant’s response to narrow-wavelength 595 nm light. These proteins play key roles in energy metabolism and redox regulation processes. Clustering analysis of the interaction network further showed two large networks (A and B), consisting of 6 DAPs, which are mainly related to energy metabolism and stress tolerance (Figure 3). Another cluster (C) was not connected to networks A and B and was composed of 3 DAPs involved in amino acid metabolism.
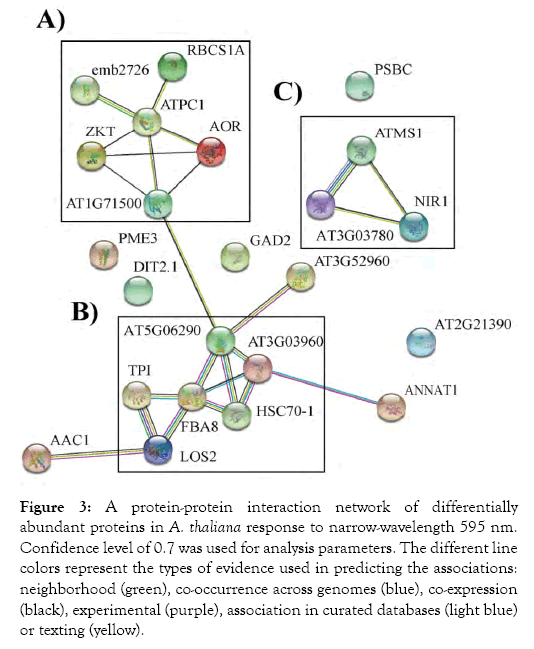
Figure 3: A protein-protein interaction network of differentially abundant proteins in A. thaliana response to narrow-wavelength 595 nm. Confidence level of 0.7 was used for analysis parameters. The different line colors represent the types of evidence used in predicting the associations: neighborhood (green), co-occurrence across genomes (blue), co-expression (black), experimental (purple), association in curated databases (light blue) or texting (yellow).
Validation of the identified differentially abundant proteins
To further confirm changes in expression level of the DAPs identified in A. thaliana response to narrow-wavelength 595 nm light, we leveraged the available labeling proteomics data where eleven-leaves plants of A. thaliana Col-0 were grown under 595 nm, and control light condition, for 5 days [32]. Proteins were extracted from the leaves of treated plants, and subsequently labeled with TMT reagents, then pooled and fractionated followed by LC-MS/ MS/MS analysis. Importantly, we observed a consistent expression pattern for 11 (79%) of upregulated proteins and 5 (56%) of downregulated proteins in this independent dataset (p-value 0.0384; permutation test).
Previous reports have noted the potential influence of narrowwavelength 595 nm light on plant stress tolerance [7,14]. We have shown that 595 nm light regulates genes associated with photosynthetic and antioxidant systems in A. thaliana Col-0 leaves (Yavari et al.). In the present proteomic analysis, a number of 595 nm light-responsive proteins were identified in A. thaliana Col-0 leaves, implying that this wavelength plays an important in the plant tolerance response.
Proteins involved in photosynthesis
Photosynthetic machinery reconfigures its components in response to changing light conditions [34]. This is true for photosystem II, and many subunits that are involved in light mediation signals [35]. Our proteomic data identified three responsive photosyntheticrelated proteins in chloroplasts that are regulated by 595 nm light. Among them, a significant increase in expression was observed for the PSII reaction center associated protein, CP43 (PSBC). This protein plays an important role in stabilizing the manganese cluster in the primary water-splitting site within the PSII complex [36,37]. This corroborates with a significant increase in CP43 transcription observed in a recent study on A. thaliana treated with 595 nm light [38]. This increase suggests that plants have a higher capacity in maintaining the stability of PSII super complex under 595 nm light, which leads to an enhanced energy transfer from the light harvesting antennae to the photosystems [39]. Consistent with this possibility, a significant decrease was observed in the relative abundance of two PSII-associated proteins, the Rieske (2Fe-2S) domain-containing protein PSB33 and MET1. Both proteins are required for PSII assembly in the plant response to light stress signals [37,40]. At the same time, a corresponding increase in expression level of GSH2, observed in a recent study on A. thaliana treated with 595 nm light, could explain further protection within plants photosynthetic machinery under 595 nm light. Overall, our data suggest a higher tolerance capacity of the PSII super-complex in plants grown under 595 nm light.
Proteins involved in carbohydrate metabolism
Plant energy metabolism and the associated proteins can rearrange to boost plant adaptations to light signaling [41,42]. Here, our analysis revealed five carbohydrate metabolism-related proteins whose expression was differentially regulated by 595 nm. Among them, two glycolysis-associated enzymes triosephosphate isomerase (TPI) and enolase 2/transcriptional activator (ENO2) were abundantly upregulated. These two enzymes are essential for plant efficient energy production [43,44]. A yeast two-hybrid study demonstrated that these two enzymes interact during glycolysis, and it was proposed that ENO2 regulates the relative abundance of TPI [44]. Importantly, alterations in relative abundance of these two proteins have been reported under abiotic/biotic stress conditions [45,46]. Data from this study further suggest that these glycolytic enzymes are involved in maintaining, as well as driving carbon metabolism in plants. There is a possibility that plants have a higher demand for ATP when maintaining metabolic homeostasis under 595 nm light condition [47]. Consistent with this prospect, our results showed an increase in the abundance of two ATP related proteins: a gamma subunit of the ATP synthase complex (ATPC1), involved in ATP production, and ADP/ATP carrier 1 (AAC1), which mediates the export of generated ATP in the mitochondrion in counter exchange with cytosolic ADP [48]. In agreement with this result, a significant increase in the expression of ATPC1 and members of the ATP synthase complex at protein level was recently reported in A. thaliana Col-0 treated with 595 nm light [32,49]. Relatedly, a recent study suggested higher chloroplast photoprotection and stress tolerance occurs in leaves of A. thaliana treated with 595 nm light, and this is due to the energy allocation from carbohydrate biosynthesis and accumulation to the production of secondary metabolites [49]. A significant downregulation of the fructose-biphosphate aldolase 8 (FBA8) could further support a restricting flux of generated energy toward the Calvin cycle [49]. Importantly, our analysis identified ATPC1 and FBA8 as core proteins in interaction networks associated with A. thaliana response to 595 nm light. Overall, our investigation suggests a high tolerance capacity in plant that is preserved by maintaining and directing the allocation of generated energy when grown under 595 nm light.
Proteins involved in amino acid metabolism
Biosynthesis of sulfur/nitrogen containing amino acids supports plant energy requirements for growth and development [50,51] and our results suggest that 595 nm light potentially impacts expression of proteins involved in amino acid metabolism. Specifically, 5-me thyltetrahydropteroyltriglutamate-homocysteine methyltransferase 1 and 2 (MS1 and MS2) involved in methionine metabolism, were significantly upregulated under 595 nm. In contrast, expression of dicarboxylate transporter 2.1 protein (DIT2-1) and ferredoxinnitrite reductase (NIR1) was downregulated in plants treated 595 nm light. DIT2-1 is involved in translocating the end product of nitrogen assimilation, glutamate, and NIR1 [52]. Further, it catalyzes the reduction of nitrite to ammonium in the second step of the nitrate-assimilation pathway [53]. This binary pattern suggests that the sulfur uptake ability of plants increased under 595 nm light, while the seedlings requirement to take up nitrate and ammonium decreased [54]. Sulfur and nitrogen-containing amino acids act as key elements in primary protein structure. Overall, our data imply a key role for these amino acids as building blocks for the synthesis of various proteins that are required for plants’ growth and developmental adjustment under 595 nm light.
Proteins involved in lipid metabolism and transport
Under high environmental stress, the thylakoid membrane disintegrates the assembled galactolipids, leading to the accumulation of free fatty acids, which are toxic to the cell [55]. In the present study, we noted a significant decrease in relative abundance of quinone oxidoreductase-like protein AT1G23740 (AOR), involved in detoxification of stromal lipid-derived reactive carbonyl species [56]. This result suggests a significant decrease in the accumulation of oxidative-modified biomolecules within chloroplasts that can support an enhanced tolerance capacity for plant chloroplast under 595 nm light. Thus, these data suggest a significant increase in thylakoid membrane adaptions in plants grown under 595 nm light.
Proteins involved in cell wall modification
Our proteomic data indicate that 595 nm light strongly influences cell wall modification, as evidenced by smaller leaves and elongated petioles; in addition, the pectinesterase/pectinesterase inhibitor 3 enzyme (PME3) was significantly abundant among DAPs. PME3 functions in the modification of cell walls, could act on cell wall plasticity and cell-to-cell adhesion [57]. The involvement of PME3 in response to 595 nm light signals suggests promoted stem elongation to position leaves at a higher position, with better chances of capturing light energy for photosynthesis. Induction of stem elongation in response to limited light energy has been previously shown [58]. Accordingly, an abundance of PME3 could be of indicative of cell organization that assists a plant’s adaptation to 595 nm light.
Proteins involved in protein synthesis, folding, and degradation
Molecular chaperones regulate protein quality control in plants [59]. Our results show that 595 nm light significantly increases the relative abundance of two chaperones, a probable mediator of RNA polymerase II transcription subunit 37e (HSC70-1) and T-complex protein 1 subunit theta (CCT8). The heat shock protein HSC70- 1 is considered a crucial protective mechanism, important in reestablishing and maintaining protein integrity in plant chloroplasts [60]. The chaperonin complex, containing CCT8, enables posttranslocational refolding of transported and targeted proteins in chloroplast [61,62]. Overall, results suggest higher adaptation for chloroplasts via protein folding, as well as strengthening protein quality control under 595 nm light.
Proteins involved in ROS signaling
In plant cells, perception of extracellular stimuli is mediated by the plasma membrane receptors [63,64]. In the present study, components in Ca2+ and ROS signaling were differentially regulated under 595 nm light when compared to control FL light. A significant increase was observed in the relative abundance of Ca2+-dependent membrane-binding protein annexin 1 (ANN1), which serves as important component in stress tolerance and maintaining Ca2+ homeostasis in plants [65,66]. Ca2+ plays an essential role in plant cells in response to environmental stimuli as a second messenger [67]. ANN1 plays a regulatory role in ROS signal transduction in a Ca2+-dependent manner in Arabidopsis [68,69]. Moreover, our results showed a high relative abundance of glutamate decarboxylase 2 (GAD2), the key enzyme involved in gamma-aminobutyric acid (GABA) biosynthesis [70]. GABA has an important function in plant developmental processes and stress responses [71,72]. Elevated GABA levels are dependent on the calmodulin domain of calcium-sensing proteins in response to cytosolic Ca2+ oscillations that occur with different environmental stresses [73]. Congruent with the potential effect of 595 nm light on ROS signaling-associated proteins, we observed that 595 nm has a significant regulatory role in activating plant antioxidant capacity, leading to higher chloroplast protection [49]. Overall, these data suggest that the calcium ion-associated signaling pathways play a pivotal role in enhancing plant adaptation, and tolerance to 595 nm light.
Proteins involved in redox signaling
Redox regulation is an essential defense response to stress conditions [74]. We observed a significant decrease in the abundance of two enzymes in the peroxiredoxin family; peroxiredoxin-2E (PRXIIE) and 2-Cys peroxiredoxin BAS1-like (AT5G06290). Under oxidative stress, PRXIIE plays an important function in the redoxsystem mechanisms that control H2O2 levels towards preserving chloroplast integrity [75] and 2-Cys peroxiredoxin BAS1-like protein functions in H2O2 detoxification [76]. Lower relative abundance of these two peroxiredoxin enzymes suggests sustaining intracellular redox homeostasis, requirements for H2O2 scavenging, in plant chloroplast under 595 nm light [77]. Overall, these data suggest that stable redox homeostasis results in higher adaptation capacity for plants growing under 595 nm light.
Here, the LC-LTQ methodology applied to Arabidopsis thaliana is the first unbiased proteomic study aimed towards the identification of molecular processes associated with a plant’s response to narrow wavelength 595 nm light. A number of proteins were identified that several of which could be involved in plant tolerance response. Although these data indicate that A. thaliana modifies its proteome upon exposure to 595 nm light, proteome coverage obtained here is low. Proteins involved in carbohydrate and amino acid metabolism were in greater abundance in plants treated with 595 nm light. Furthermore, molecular chaperones and PSII-associated membrane proteins showed significant changes in abundance. The comparative phenotype analysis suggests that 595 nm light also affects the cell wall proteome.
NY and ML designed the experiments. NY performed the experiments and analyzed the data. ML contributed reagents/ materials/analysis tools and supervised the project. NY and ML wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC; grant number RGPIN 355743-13, CRDPJ418919-11).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Citation: Yavari N, Lefsrud MG (2019) Proteomic Analysis Provides Insight into Arabidopsis thaliana’s Response to Narrow-Wavelength 595 nm Light. J Proteomics Bioinform 12:507. doi: 10.35248/0974-276X.19.12.507
Received: 25-Nov-2019 Accepted: 23-Dec-2019 Published: 30-Dec-2019 , DOI: 10.35248/0974-276X.19.12.507
Copyright: © 2019 Yavari N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.