Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2021)Volume 14, Issue 3
Huanglongbing (HLB or Citrus Greening Disease) is a destructive citrus disease that has impacted the U.S. citrus industry as well as global citrus industry for many years. There is yet to be a practical cure for HLB. Currently, there are only temporary and costly solutions of applying insecticides to target the psyllid vector and burning down the infected trees. Here we investigated the protein and phosphoprotein differences between control (healthy) and diseased citrus leaves to identify potential biomarkers essential towards fighting HLB within the trees themselves. Proteins were extracted from control and diseased leaves, followed by trypsin digestion, phosphopeptide enrichment and label-free quantitative proteomics. In total, we identified 1539 proteins and 278 phosphoproteins, among which 63 proteins and 23 phosphoproteins exhibited significant changes between control and diseased samples. Proteins in response to stimulus were increased at both total protein level and phosphorylation level. Interestingly, proteins related to membranes and complexes were decreased, but increased at phosphorylation level. Several differential phosphoproteins (e.g., a nuclear pore complex protein and a glutathione-S-transferase) could play important roles in battling the devastating disease HLB, one that we can hopefully conquer in the near future.
Huanglongbing; Citrus sinensis; Proteomics; Phosphoproteomics; Molecular mechanisms
Citrus greening is a deadly disease that has devastated global citrus communities for almost a century [1,2]. Citrus greening is known by many epithets around the globe, including Huanglongbing (HLB) (yellow dragon disease), Citrus vein phloem degeneration, and yellow shoot disease to name a few. Each of these names refer to the pathogenic, gram-negative α-proteobacteria known as Candidatus Liberbacter (abbreviated C. Liber) [3]. There are three known species of the C. Liber genus that infect citrus, aptly named depending on the bacterium’s origin: C. Liber. asiaticus (CLas), C. Liber africanus, and C. Liber americanus [1,4]. The first documented case of the disease was reported in 1919 in China [1]. Later, in 1937 an African variant was reported in South Africa, and it finally appeared in the United States in the state of Florida in August 2005 [1,5]. In 2016, 90% of Florida’s citrus acres were infected with HLB, with approximately 80% of the citrus population infected with the disease [6]. HLB was responsible for 72.2% reduction (from 8.0 to 2.2 billion tons) in the yield of oranges for processing in the United States during the past decade [5].
C. Liber is transmitted through two main psyllid vectors: The Asian citrus psyllid, Diaphorina citri, and the African citrus psyllid, Trioza erytreae. The psyllids transfer the bacteria after encountering it in the phloem of an infected plant. As the bacterium is known only able to thrive in the phloem of citrus plants and in the microbiome of the psyllid vector, it has been near impossible to culture and study C. Liber in vitro. C. Liber infected citrus trees display a variety of symptoms, including accumulation of starch, chlorosis of the leaves, and decrease in growth [1,7,8]. Fruits produced by the plants are irregularly shaped, colored and taste sour and bitter, making them inferior for consumption and marketing. Eventually, the damages incurred by the plant result in its inevitable death within 5 years. This disease has cost the U.S. agriculture industry an approximate $4.5 billion in total damages and losses [9]. Many methods have been proposed to combat the pathogen, such as controlling the psyllid vectors through the use of pesticides, creating disease-free nursery through the use of sterilized propagating material, and by simply burning the infected trees [1,10]. However, as there can be a latency period between the time of infection and the time of detection, most of the methods are simply inefficient and unsustainable to quarantine and eradicate the disease [11].
Recent metabolomics, transcriptomics, and proteomics efforts have provided a few clues as to how the pathogen interacts with its vector and host. In metabolomics, work has been done in identifying citrus response to C. Liber infections, with a recent study documenting increases of seven amino acids (glycine, isoleucine, phenylalanine, proline, serine, threonine, and tryptophan) and five organic acids (benzoic acid, citric acid, fumaric acid, salicylic acid (SA), and succinic acid) after infection with CLas, indicating that these amino acids and organic acids may play a role in the defense of citrus plants, particularly in the activation of SA [12]. In transcriptomics, several studies found that genes related to photosynthesis (mainly in the light reaction) and carbohydrate metabolisms were down-regulated in HLB infected plants [13-15]. Conversely, genes related to oxidation-reduction (redox), transport, starch biosynthesis, and sucrose metabolism are up-regulated [13-17]. A recent study has identified transcripts of constitutive disease resistance genes (CDR) involved in HLB resistance [18]. However, it is well known that transcripts do not correlate well with protein levels and cannot predict protein post-translational modifications (PTMs) [19]. Thus, proteomic analyses must be conducted to accurately decode the mechanisms of HLB.
In proteomics, multiple proteins were identified in HLB infected citrus. They include phloem-specific lectin PP2-like proteins, miraculin-like proteins, Cu/Zn superoxide dismutase, copper ion binding protein, germin-like proteins, subtilisin-like proteins, and serine carboxypeptidase-like 40 proteins [11,20-22]. The phloem-specific lectin PP2-like proteins are used for carbohydrate binding and accumulation [23]. The other proteins (miraculin- like proteins, Cu/Zn superoxide dismutase, and copper ion binding protein, germin-like proteins, subtilisin-like proteins, and serine carboxypeptidase-like 40 proteins) are all found in defense pathways and stress signaling pathways [24-27]. A recent study has also shown that HLB defense in citrus leaves was correlated with an induction of proteins involved in detoxification, e.g., glutathione S-transferases (GSTs) [28].
Although these transcriptomics and proteomics studies have identified some key differences between healthy and HLB infected citrus, to get a clear picture of the effects of HLB we should consider the various post-translational modifications, especially protein phosphorylation. By studying phosphoproteins we can identify key proteins involved in signal transduction pathways and metabolic regulations, which are critical for citrus defense. In this study, we report changes in both the total proteome and phosphoproteome of CLas-infected and control Citrus sinensis leaves. Our aim is to identify potential phosphoproteins needed in the citrus’ defense against HLB in hopes to prevent further catastrophic damage by C. Liber species to citrus and to other potential hosts.
Growth conditions of tree seedlings
Trees were acquired from Phillip Rucks Citrus Nursery from Frostproof, FL, USA. The seedlings are of the Valencia Orange variety (Citrus sinensis) on Kuharski rootstock. Plants were grown in a Fafard Citrus Mix soil from Sungro Horticulture (Agawam, MA, USA) in a greenhouse with a controlled temperature of 28°C and humidity of 74%. The seedlings were watered once every three days for one minute with a flow-regulated drip line from Jain Irrigation Systems Ltd (Fresno, USA) with a flow rate of 2 L/min. A fertilizer composed up of 20% nitrogen, 10% phosphate, 20% potash, 0.15% water magnesium, 0.0125% boron, 0.0125% chelated copper, 0.05% chelated iron, 0.025% chelated manganese, 0.005% molybdenum, and 0.025% chelated Zinc was dissolved in water to produce a 5% stock solution. It was then diluted 100 times in water to achieve a final nitrogen concentration of 100 ppm and applied every other week. HLB infected trees were generated as previously described [29].
Leaf protein extraction using a phenol method
Two grams of fresh leaves (as one biological replicate) of healthy and infected leaves were ground in liquid nitrogen in a mortar and pestle. A phenol method [30] was used to extract the proteins from four biological replicates of the control (healthy) leaves and four biological replicates of the treatment (infected) leaves. Briefly, 5.0 mL of Tris pH 8.8 buffered phenol and 5.0 mL of extraction media (0.1 M Tris-HCl pH 8.8, 10 mM EDTA, 0.4% 2-mercaptoethanol, and 0.9 M sucrose) was added to the leaf powder and continued grinding for 30 seconds. The samples were transferred to Falcon tubes and agitated them at 4oC for 30 min. After centrifugation at 5000 g and 4oC for 10 min, the phenol phase was removed, and the aqueous phase was back-extracted with 5 mL phenol and 5 mL extraction buffer). The second extraction was combined with the first extraction. Then five volumes of 0.1 M ammonium acetate in 100% methanol was used to precipitate the phenol extracted proteins overnight at -20oC. Afterwards, the precipitate was collected through centrifugation at 20,000 g, 4oC for 20 min. The samples were washed five times each, two times with 0.1 M ammonium acetate in methanol, two times with ice cold 80% acetone and finally once with cold 70% ethanol (-20oC). The final pellet was solubilized in 6 M urea and 0.1% SDS with agitation at room temperature for 1 h.
Protein assay, gel electrophoresis, and trypsin digestion
A Bradford assay (BioRad, Hercules, CA, USA) and 1-D SDS gel electrophoresis were used to evaluate the quantity and quality of the proteins. The samples were diluted six times in 50 mM ammonium bicarbonate, pH 8.5, and then reduced by 10 mM tris(2-carboxyethyl)-phosphine at room temperature for 1 h, followed by alkylation by 20 mM iodoacetamide in the dark for 30 min. Trypsin (Promega, Fitchburg, WI, USA) was added (w/w for enzyme : sample=1 : 100) for digestion overnight at 37°C. After trypsin digestion, each sample was split into two groups: one for phosphopeptide enrichment (90 μg) and the other for total proteomic analysis (10 μg).
Phosphopeptide enrichment and ZipTip clean up
Phosphopeptides were enriched with the NuTip (TiO3/ZrO2, Glygen, Columbia, MD, USA) according to manufacture instructions. Briefly, the protein digest was lyophilized to dryness and then solubilized in 10 μl of sample buffer (3% acetonitrile (ACN), 0.1% acetic acid +0.01% trifluoroacetic acid (TFA)) and 10 μl of binding solution (80% ACN, 5% TFA). The peptides were loaded into the NuTip by aspired and expelled 100 times. The unbound peptides were then rinsed off using 200 μl of washing solution (80% ACN, 1% TFA with pH less than 3). Phosphopeptides were eluted by aspiring and expelling 50 times with an elution solution (3% NH4OH in water, pH=11). Finally, the eluted peptides were acidified using 2 μl of formic acid. The samples were dried using a speedvac and stored at -20°C. The other 10% of peptides were cleaned up using ZipTip solid phase extraction according the manufacturer’s instructions (MilliporeSigma, St. Louis, MO, USA).
LC-MS/MS analyses of total peptides and phosphopeptides
Peptides derived from the protein samples were resuspended in 0.1% formic acid for mass spectrometric analysis. The bottom- up proteomics data acquisition was performed on an EASY-nLC 1200 ultra-performance liquid chromatography system (Thermo Scientific, San Jose, CA, USA) connected to an Orbitrap Fusion Tribrid instrument equipped with a nano-electrospray source (Thermo Scientific, San Jose, CA, USA). The peptide samples were loaded to a C18 trapping column (75 μm i.d. × 2 cm, Acclaim PepMap® 100 particles with 3 μm size and 100 Å pores) and then eluted using a C18 analytical column (75 μm i.d. × 25 cm, 2 μm particles with 100 Å pore size). The flow rate was set at 300 nL/ minute with solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid and 99.9% acetonitrile) as the mobile phases. Separation was conducted using the following gradient: 2% of B over 0-3 min; 2%-40 % of B over 3-150 min, 40%-98 % of B over 150-151 min, and isocratic at 98% of B over 151-166 min, and then from 98–2% of B from 166–167 min. The equilibration at 2% B is from 167 to 180 min. The full MS1 scan (m/z 35-2000) was performed on the Orbitrap with a resolution of 120,000 at m/z 400. The automatic gain control (AGC) target is 2e5 with 50 ms as the maximum injection time. Monoisotopic precursor selection (MIPS) was enforced to filter for peptides. Peptides bearing +2 to +6 charges were selected with an intensity threshold of 1e4. Dynamic exclusion of 15 s was used to prevent resampling the high abundance peptides. Top speed method was used for data dependent acquisition within a cycle of 3 s. The MS/MS was carried out in the ion trap, with a quadrupole isolation window of 1.3 Da. Fragmentation of the selected peptides by collision induced dissociation (CID) was done at 35% of normalized collision energy. For peptides with charges between +4 and +6, electron transfer dissociation (ETD) was activated. The MS2 spectra were detected in the linear ion trap with the AGC target as 1e4 and the maximum injection time as 35 ms. The data generated in this study have been submitted to Massive repository and can be accessed via MSV000084179.
Label-free proteomics data analysis
The raw data files were processed with the Proteome Discoverer (PD) software (version 2.2, Thermo Scientific, San Jose, CA, USA) using a citrus database containing entries from Citrus sinensis and C. clementina (www.citrusgenomedb.org). The precursor mass tolerance was set to 10 ppm with a fragment mass tolerance of 0.5 Da. Trypsin was selected as the enzyme and the maximum missed cleavage of 1 was set. Carbamidomethylation (C) was set as fixed modification with variable modifications of oxidation (M) and acetylation (N-term). The PD processing workflow starts with loading raw files using the “Spectrum Files” node, followed by a “Spectrum Selector” for optionally filtering spectra based on various criteria. Then the workflow branches: MS1 peptide features are quantified in all runs by “LFQProfiler FF”, and MS2 spectra are identified using the native PD node Sequest HT. Subsequently, peptide identifications are validated using Percolator. The consensus workflow is used to combine the individual processing results. It starts with the obligatory “MSF Files” node for loading the processing results stored in a Thermo MSF file. “LFQProfiler” then exports peptide identifications and map onto their corresponding quantified features. Statistical data analyses were performed in R (version 3.5.1) to determine different abundance proteins. A p<0.05 and two-fold change cutoff was applied to determine differential proteins and phosphoproteins. Arabidopsis thalianaIDs for the identified protein sequences were generated by a local blastp algorithm against TAIR10 (20190711) with a e-value cutoff at 0.01. Gene ontology (GO) annotation was identified by the Arabidopsis IDs and WEGO [31] was used for GO visualization. Cytoscape was used for differential abundance protein-protein interaction networks using STRING protein database with confidence cutoff at 0.8 and maximum additional interactors at 40. Mapman [32] was used for protein functional annotation in metabolic pathways using the A. thaliana IDs and protein change ratios (log2(treatment/ control)).
Multi-omics data analysis
To correlate proteomics and phosphoproteomics data acquired in this study with transcriptomics, we downloaded GSE29633 [11]as the mRNA data set for comparison to the proteomics and phosphoproteomics data. The rationale for selecting this data set is that they used the Affymetrix GeneChip Citrus Genome Array. The data from this chip can be directly matched to our proteomics data through the same accession numbers. For multi-omic correlation, the changing trends of the significantly differential proteins, phosphoproteins and transcripts were compared.
Phenotype of healthy and CLas infected leaves and protein quality
Sweet oranges of the species C. sinensis are known to be HLB- sensitive [14]. The phenotypes of healthy and CLas infected C. sinensis leaves matched well those from previous literature [1]. The top leaf, the control, is shown to be healthy and uninfected, as seen by its uniform color and smooth texture in Figure 1A. The bottom leaf, the treatment, is shown to be infected with CLas as the leaf is seen to have dark, blotchy mottling, a little bit of yellowing, and a thicker/leathery texture. Leaves from four control seedlings and those from four diseased seedlings were used as biological replicates for proteomics and phosphoproteomics experiments. After SDS gel electrophoresis and Commassie blue staining, the proteins from different samples fell within a 14 kDa and 200 kDa range. The quality was acceptable and the amounts appeared equal among the different replicate samples (Figure 1B).
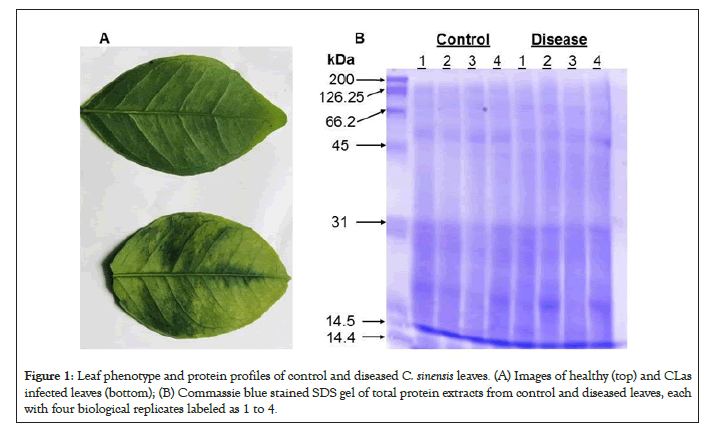
Figure 1: Leaf phenotype and protein profiles of control and diseased C. sinensis leaves. (A) Images of healthy (top) and CLas infected leaves (bottom); (B) Commassie blue stained SDS gel of total protein extracts from control and diseased leaves, each with four biological replicates labeled as 1 to 4.
Proteomic and phosphoproteomic overview of healthy and CLas infected leaves
Using label-free quantitative proteomics of total proteins, we observed 1539 non-redundant proteins, 63 of which were differentially expressed proteins (DEPs) between healthy and infected leaves as seen in Figure 2. Using enriched phosphopeptides, we identified a total of 278 phosphoproteins, 23 of which were DEPs between healthy and infected leaves (Figure 2). Based on GO functional Classification shown in Figure 3, proteins in response to stimulus increased in both total and phosphoproteome groups, by 27% and 25%, respectively. Proteins with catalytic activity were also increased in both groups, by 21% and 11%, respectively. In contrast, proteins related to organelles, organelle parts and binding all decreased in both groups by 10% and 2%, 15% and 3%, and by 22% and 19% respectively. Proteins related to membranes decreased by 7% in the total proteome group, but increased by 25% in the phosphoproteome group. Proteins related complex decreased by 6% in the total proteome group, but increased by 44% in the phosphoproteome group. These results not only revealed biological processes and molecular functions altered by the CLas infection, but also highlight the importance of conducting phophosphoproteomics in addition to total proteomics. For examples, those proteins with decreased levels but increased phosphorylation may function in the kinase phosphorylation cascade, and thus may play important roles in signal transduction and defense responses.
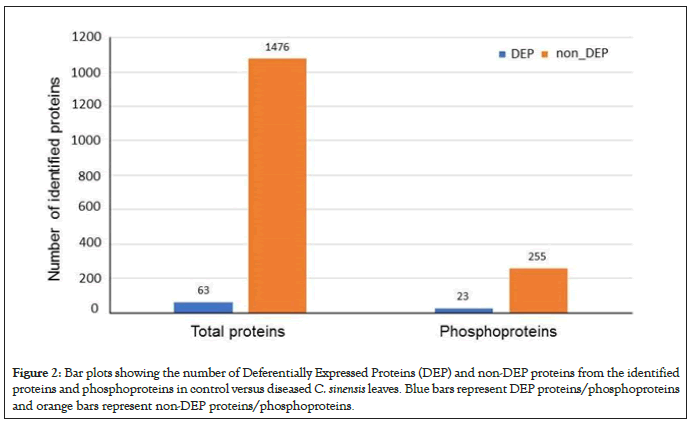
Figure 2: Bar plots showing the number of Deferentially Expressed Proteins (DEP) and non-DEP proteins from the identified proteins and phosphoproteins in control versus diseased C. sinensis leaves. Blue bars represent DEP proteins/phosphoproteins and orange bars represent non-DEP proteins/phosphoproteins.
Figure 3: GO enrichment of DEPs between control and diseased C. sinensis leaves (A) GO enrichment of total protein DEPs; (B) GO enrichment of phosphoprotein DEPs. Red bars represent increased proteins after CLas infection; blue bars represent decreased proteins after CLas infection.
CLas infection-induced protein level changes
As shown in Figure 2, 63 DEPs showed total protein level changes in response to the CLas infection. Here we discuss a few interesting DEPs. One was a chloroplastic ferritin-3, which is used in iron storage and exhibits ferroxidase activity [33]. Chloroplast is one of the major iron sinks in the plant cell, and thus it is prone to oxidative damage [34,35]. In our samples, the ferritin-3 was increased by 10.2 fold. With ferroxidase activity, it could oxidize iron II to iron III to prevent hydroxyl radical production and damage associated with the CLas infection [36]. However, the overall antioxidant activity was decreased (Figures 3A and 3B). This could be due to a multitude of factors, including the well- known suppression of host anti-oxidative defense by pathogens [37]. Another factor could be the impaired nutrient absorption in the course of HLB development [22]. Thus, a lack of iron may feedback regulate the expression of ferritin-3, as observed here. These possibilities present exciting future research directions toward understanding the citrus greening disease. Protein DJ-1 homolog B-like was also increased (by 5.2 fold). It very interesting to find this protein as mutation to its human homolog DJ-1 is known to be associated with Parkinson’s disease and premature cell death. When the protein was overexpressed in Arabidopsis, the plants showed enhanced protection against environmental stresses through activation of superoxide dismutase [38]. The molecular mechanism underlying the activation is not known. In addition, glutathione-S-transferases (GST) were increased (Supplemental Table 1). For example, the most predominant GST23 showed a five-fold increase. GSTs have been shown to be crucial in cellular detoxification during oxidative stress response [35,39]. Previous research has shown that HLB indeed causes oxidative stress, and GSTs were proposed to be a likely component for relieving the oxidative stress [40]. Thus, our observed increase in GSTs supports the hypothesis in the previous study.
Another DEP showing a 6.5 fold increase after the CLas infection was 3-isopropylmalate dehydratase small subunit 3-like (SSU3), which is involved in leucine biosynthesis and female gametophyte development [41-43]. The increase of SSU3 correlates with the slight enrichment of the branch-chain amino acid pathway (Figure 4A). It is possible that SSU3 has other functions to contribute to the HLB phenotype, together with blocked phloem transport [22]. Interestingly, proteins related to auxin and jasmonate (JA) biosynthesis were over-represented after CLas attack (Figure 5A). Given that JA and salicylic acid (SA) are antagonistic hormones [44,45], it was quite surprising to see increases in JA-related proteins (Figure 5A). The biological implications of this result need to be further investigated.
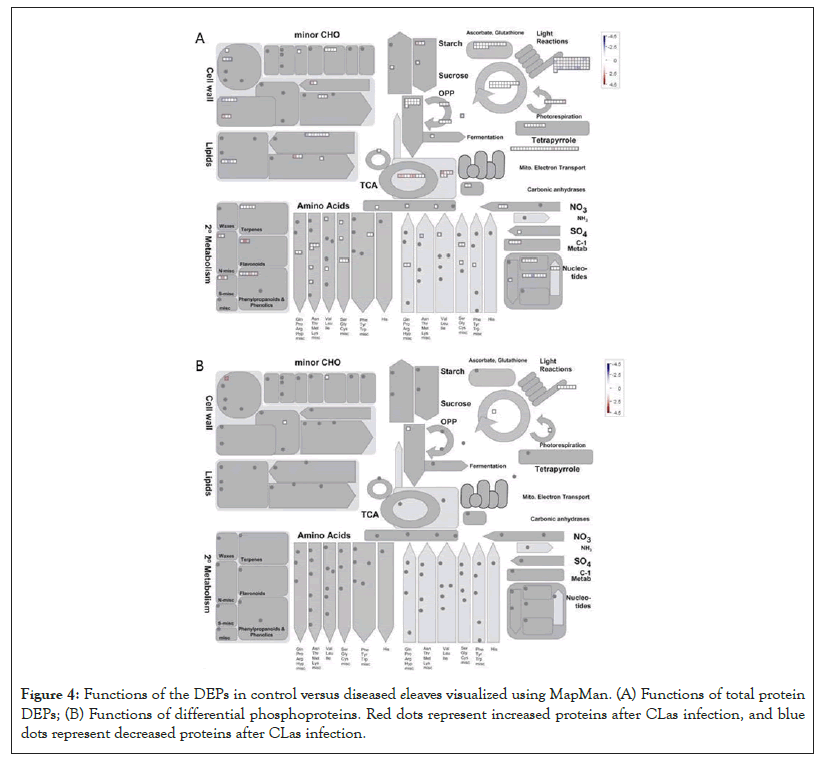
Figure 4: Functions of the DEPs in control versus diseased eleaves visualized using MapMan. (A) Functions of total protein DEPs; (B) Functions of differential phosphoproteins. Red dots represent increased proteins after CLas infection, and blue dots represent decreased proteins after CLas infection.
As to the decreased proteins, one was translation initiation factor IF3-4 with a 8.6 fold decrease after the CLas infection. IF3 has been shown to regulate chloroplastic gene translation initiation [46]. We hypothesize that the decrease of IF3 in CLas infected plants may account for the blotchy mottling and yellowing of the citrus leaves. This decrease of IF3 correlates to the decrease of photosynthetic light reactions shown in Figure 4A. IF3 has also been shown to play a major role in auxin homeostasis [47]. However, the overall increase in the auxin biosynthesis after the CLas infection may promote the expression of expansins, breakdown of the cell wall, and thus HLB development [48,49]. Another protein that was decreased was peroxiredoxin Q (PrxQ). It catalyzes the conversion of hydrogen peroxide and organic hydroperoxides into water. Thus, PrxQ plays a critical role in protecting the cells from oxidative stressors, especially the photosystem II complexes in the chloroplasts. It has also been observed that PrxQ deficient A. thaliana mutants had a lower sensitivity to oxidants [50]. Thus, the decrease in PrxQ that we observed is likely to contribute to an overall decrease in activity in redox/antioxidant responses shown in Figure 5A.
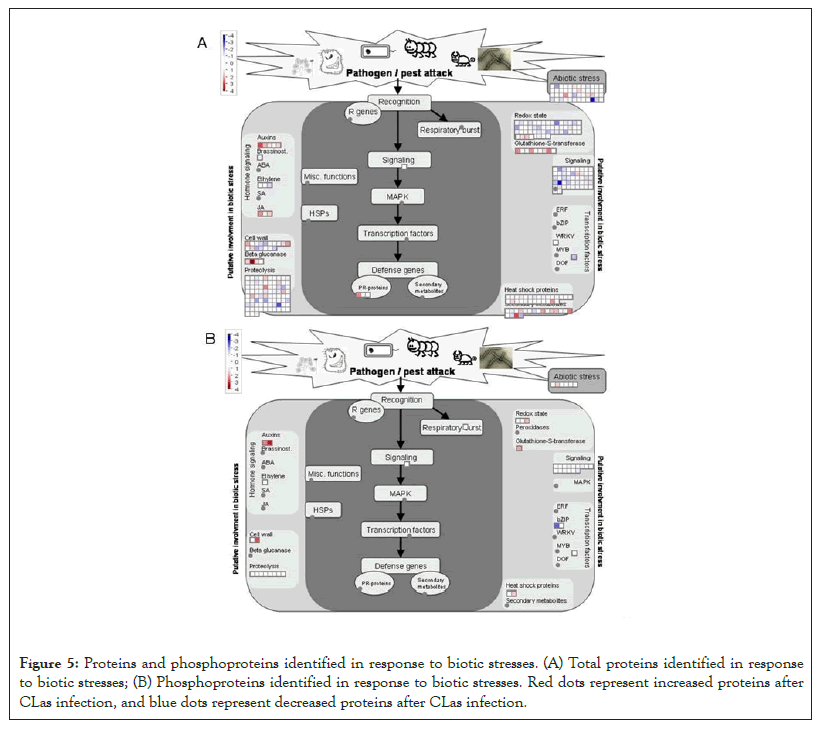
Figure 5: Proteins and phosphoproteins identified in response to biotic stresses. (A) Total proteins identified in response to biotic stresses; (B) Phosphoproteins identified in response to biotic stresses. Red dots represent increased proteins after CLas infection, and blue dots represent decreased proteins after CLas infection.
CLas infection-induced protein phosphorylation changes
In total, 278 phosphoproteins were identified. Among them, 23 phospho-proteins were observed to have increased or decreased in abundance (Figure 2, Supplemental Table 2). One such protein that increased in phosphorylation by 4.4 fold was nuclear pore complex (NPC) protein NUP98A. In humans and yeasts, NPCs are crucial in protein and RNA transport as well as nuclear pore reconstruction following mitosis. In plants, there has been limited research done on NPCs. In Arabidopsis, NPCs have been shown to be involved in pathogen interactions and hormone signaling [51]. How phosphorylation regulates NPC functions and what role it plays in HLB disease development are interesting questions for future studies. Another protein GSTL3 showed increased phosphorylation by 4.1 fold. A previous work using 2D gel to study SA induced plant GSTs did not identify phosphorylation of GSTs [52]. In this study, detection of GSTL3 phosphorylation is significant as GSTL3 is part of the lambda CLass of GSTs (GSTLs), which catalyze the glutathione-dependent reduction of S-glutathionyl-quercetin to quercetin under stress conditions [53]. Flavonoids are important in plant pathogen defense due to their antioxidative properties and inhibition of pathogen enzymes [54]. An increase in the phosphorylated GSTL3 (Figure 5B) indicates that during CLas infection, C. sinensis increased GSTL3 activity to maintain and increase flavonoids for combating ROS and CLas bacterium. An increase in the phosphorylation of GSTL3 could also explain the enrichment of the phosphorylation pathways of redox and auxin as seen in Figure 5B. Previous literature has shown that flavonoids tend to inhibit auxin pathways, facilitating pathogen defense by protecting the natural pathogen barrier cell walls [54]. It is possible that the increase of flavonoids is a response to the increase of auxins induced by CLas during the infection. However, further research should be conducted to reach a conclusive answer.
As to decreased phosphorylation, the transport-related proteins stand out (Figure 3B). For example, phosphorylation of a SEC16B isoform X1 showed a 2.3 fold decrease. SEC16B plays a role in the transport of proteins between endoplasmic reticulum (ER) and Golgi apparatus [55]. In contrast, the total level changes of transport-related proteins were highly enriched (Figure 3A), but phosphorylation was decreased (Figure 3B), indicating phosphorylation may not be important in regulating the transport activity. A signaling-related VQ motif-containing protein (VQP) 4 was also decreased in phosphorylation. Previous research has shown that VQPs are phosphorylated by mitogen-activated protein kinases (MAPKs), specifically MPK3 and MPK6 [56]. When plants expressing these MPK3/6-targeted VQPs (MVQs) were treated with flagellin-derived peptide flg22, the MVQs were destabilized. As MVQs are negative regulators of WRKY-mediated defense gene expression, a destabilization of these MVQs allowed for up- regulation of defense gene expression [56]. In our samples, we observed a two-fold decrease in the phosphorylation of VQPs in the CLas infected leaves compared to healthy leaves. This indicates that phosphorylation of the MVQs decreased after the CLas infection, thus MVQs became stable, leading to decreased plant defense response (Figures 3B and 5B) and the disease phenotype (Figure 1).
Correlation between transcript level, protein level and phosphoprotein level
The overall correlation of differential molecules in the transcriptome, proteome and phosphoproteome is relatively low (Supplemental Table 3), despite higher matching in the identified molecules at the different levels. This result can be attributed to the incomplete coverage of each omic technology, as well as the different levels of regulations. It highlights the complementary nature of each omic analysis, and the importance of conducting multi-omics [19,57,58]. For example, transcription of a GST L3 increased, but did not lead to any changes at the protein level. However, its phosphorylation was increased after the CLas infection (Supplemental Table 3). This result is consistent with a previous report of GST L3 changes under high temperature stress [59]. The biological implication of the GST L3 phosphorylation needs to be further studied. For all the differential proteins, their corresponding transcript levels either did not change significantly or were not detected. This discordance between the protein level and RNA level is well-known, and may be attributed to PTMs, different dynamics and stability of mRNAs and proteins [19]. It is not surprising that the phosphorylation level changes exhibited the least correlation with the transcript changes, since transcript levels cannot predict the protein PTMs. On the other hand, the decrease in phosphorylation of a 28 kDa ribonucleoprotein may be attributed to the decrease of its protein level (Supplemental Table 3), again highlighting the utility and importance of multi-omics.
The study aimed to detect and measure differentially expressed proteins and phosphoproteins (DEPs) between healthy C. sinensis plants and those infected by CLas. Using LC-MS/MS-based label- free quantitative proteomics and phosphopeptide enrichment, we were able to identify 1539 proteins and 278 phosphoproteins. Among them, 63 proteins and 23 phosphoproteins were DEPs. In the total protein group, multiple proteins related to stress, redox and hormone pathways appeared to play an important role in CLas defense. In the phosphoprotein group, we discovered interesting phosphorylation changes in several proteins, such as the nuclear pore complex, GSTL3, SEC16B and VQ motif protein 4 (VQP4). The increase of phosphorylation of GSTL3 may regulate cellular flavonoid contents to alleviate oxidative stress during pathogen infection. We also noted the decreased phosphorylation of VQP4 in the MVQ pathway, potentially contributing to the HLB phenotype. Our analysis of the enrichment of phosphoprotein pathways and especially the phosphoproteins (e.g., VQP4 in MPK signaling) may facilitate the development of novel approaches to combating CLas infection of citrus plants in the future.
The authors would like to thank Drs. Lei Pan, Craig Dufresne and Wei Zhu for advice and assistance with the experiments.
The authors declare no conflict of interest.
Citation: Chen D, Guan Q, Cao B, Chen S (2021) Quantitative Phosphoproteomics of Sweet Orange (Citrus sinensis) Infected by Candidatus Liberibacter. J Proteomics Bioinform. 14:528.
Received: 26-Jan-2021 Accepted: 09-Feb-2021 Published: 16-Feb-2021 , DOI: 10.35248/0974-276X.21.14.528
Copyright: © 2021 Chen D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.