Research Article - (2024)Volume 6, Issue 2
The purpose of this research is to understand the biological regulation mechanism of 5-hydroxymethylcytosine (5hmC) formation in distinct murine brain regions during aging. Numerous studies demonstrate the enrichment of 5hmC in brain. Being the intermediate in Deoxyribonucleic Acid (DNA), demethylation 5hmC it is also regulated by certain chromatin structures. As there is an unknown biological mechanism in brain we investigated the dynamics of H3K4me3 and H3K27me3 modifications for Ten-eleven translocation (Tet) gene expressions and for the known quantitative 5hmC levels by focusing distinct developmental status (embryonic day 16, day of birth, day 7, 15, 30 and 120 after birth) of frontal and cerebellar cortex in murine brain. In this research, we summarize the findings in availability of Tet gene and of 5hmC, and their dependence on transcriptional regulation at H3K4 and H3K27. Furthermore, we discuss future challenges and directions.
Gene expression; DNA methylation; histone modification; polymerase chain reaction; epigenetic regulation
The major mechanisms of epigenetic regulation are DNA methylations and histone modifications [1,2]. Methylation of DNA is a process by which at the 5-position (5mC) a methyl group is added to cytosine. It is essential for several cellular processes [3,4], and it is modulated by three members of the Teneleven translocation (Tet1, Tet2, Tet3) family. They oxidize into 5-hydroxymethylcytosine (5hmC), further into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) [5-9]. 5hmC, widely accepted as the sixth base in mammalian genome, is known for its region and age dependent distribution in brain [8,10]. Interestingly, Tet proteins are responsible for decreased expressions in frontal and cerebellar cortex during aging [11], collectively suggesting the initiation of active or passive DNA demethylation [12,13].
Histone modifications have been shown to regulate DNA methylation. The most extensively studied histone modification is methylation, especially of histone H3 lysine 4 (H3K4) and lysine 27 (H3K27) [14]. While H3K4 trimethylation (H3K4me3) is an active mark for transcription, H3K27 trimethylation (H3K27me3) is generally associated with transcriptional repression [15,16].
The aim of this study is to understand the biological regulation mechanism of 5-hydroxymethylcytosine (5hmC) formation in distinct murine brain regions during aging. We investigated the dynamics of H3K4me3 and H3K27me3 modifications for the Tet gene expressions and the still known quantitative 5hmC levels by focusing distinct developmental status (embryonic day 16, day of birth, day 7, 15, 30 and 120 after birth) in the frontal and cerebellar cortex of murine brain.
Sample data
Healthy murine brain tissues (strain C57Bl/6) were selected, female and male mice were used. The samples were assigned to six age groups beginning at embryonic day 16 of development, then day 0, 7, 15, 30 and 120 after birth. Five cases per group were used. The removed brains were stored at -80°C. Frontal and cerebellar cortex were selected as target regions.
Extraction of RNA and reverse transcription reaction
Ribonucleic Acid (RNA) extraction was performed using the RNeasy micro kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol including a DNAse digestion step. We diluted the extracted RNA in 15 lL RNAse-free water. Reverse transcription reaction of RNA into cDNA was performed using the Superscript Vilo cDNA synthesis kit (life technologies, Darmstadt, Germany) and up to 2.5 lg of extracted RNA (500 ng/lL) according to the manufacturer’s instructions.
Quantification of gene expression using real-time PCR
For real-time Polymerase Chain Reaction (PCR), equal amounts of cDNA were used with Power SYBR green PCR master mix (Life Technologies, Darmstadt, Germany) on a LightCycler 480 II System (Roche, Basel, Switzerland) according to the manufacturer’s protocols. The gene expression levels were calculated according to the comparative CT-method (DDCTmethod). Expression levels were normalized to Gapdh, Hprt, Ipo8 and Tbp [11].
The primer sequences for Tet1 gene: Forward 5'- GAAGCACCGTGGTGTGTAC-3', reverse 5'- GAACAGGCTCAGTAAAACGTAG-3'; for Tet2 gene: Forward 5'- GGCAGTACAGTGGTGGTCA-3', reverse 5'- CTTGGCTTTCTTGGGCTCC-3'; for Tet3 gene: Forward 5'- GTGCACTGTGGTCTGCAC-3', reverse 5'- CTCTGGGGAATGCTGTGAG-3'; for Gapdh: Forward 5'- GAGAAACCTGCCAAGTATGATG-3', reverse 5'- CTTGACAAAGTTGTCATTGAGAGC-3'.
Chromatin immunoprecipitation assay
Frozen brain tissue was performed on ice beginning by homogenizing in 250 μl PBS. Cross-linking was done by incubation in 1% formaldehyde at 37% for 10 minutes, followed by adding glycine to a final concentration of 0.125 M. Genomic DNA was sheared to 200-500 bp using EpiSheary™ Probe Sonicator (Active Motif, California, USA: Catalog no. 53051) at optimal shearing conditions. The remaining steps based on the protocol MAGnify™ Chromatin Immunoprecipitation System (invitrogen, California, USA: Catalog no. 492024). Diluting sheared chromatin by adding Proteinase K followed by binding to Dynabeads (Dynabeads™ Protein G Immunoprecipitation Kit; invitrogen, California, USA: Catalog no. 10007D). Diluting sheared chromatin to 1:10 by adding Proteinase K. Lysates were then separated to pellet debris by binding to an Antibody- Dynabead complex which includes the primary antibody (anti- H3K4me3, anti-H3K27me3) (Active Motif, California, USA: Catalog no. 39060, 39055) or the rabbit IgG antibody (control) coupled to Dynabeads. Immunoprecipitation was carried out on rotator at 4°C overnight. Then the bound chromatin was washed three times with 100 μl IP buffer 1 and two times with IP buffer 2. Cross-linking was reverted by adding proteinase K and incubating by heating at 55°C for 15 minutes, followed by a further boiling at 65°C for 15 minutes. Finally, un-crosslinked DNA was purified using magnetic beads and isolated by incubation for 20 minutes at 55°C. The resulting immunoprecipitated DNA was quantified with the Qubit fluorometer by using the Qubit™ dsDNA HS assay kit (invitrogen, California, USA: Catalog no. Q32851) and stored at -20°C until subjecting quantitative real-time PCR. All primers were synthetized by Eurofins MWG Operon despite Gapdh-2 and were performed specific for 17-28 bp. Gapdh-2 amplification was selected for data normalization.
Analysis of qRT-PCR
The regulation of all Tet genes at genomic DNA with the modifications H3K4me3 and H3K27me3 was investigated using the quantitative Real-Time PCR (qRT-PCR) technique. qRTPCR was performed on the LightCycler 480 II (Roche, Germany) using SYBR™ Gren (Applied Biosystems, USA: Catalog no. A25741). Selected reference gene was Gapdh-2 (Active Motif, California, USA: Catalog no. 71018). The primer sequences for the Tet1 gene: Forward 5'- CCAGCTCACCCTAAACTGC-3', reverse 5'- CTGGAAAGTTTGTCCAAGGATTG-3'; for Tet2 gene: Forward 5'-CCGTCAAGAGCGAGGAAAG-3', reverse 5'- GGTGGACTGCGAGGCTG-3'; for Tet3 gene: Forward 5'- GCAAGCCACTTTAGAACTTGC-3', reverse 5'- CATCACAGGTCATTTTGTAAAATAAAAG-3'. The experiment was repeated in triplicates with a final volume of 10 μl. Each sample contained 3 μl of gDNA, 2 μl of primer pair and 5 μl of SYBR Green (Applied Biosystems, Germany) in PCR 96- Well-Plate (Catalog no. 712282) (Biozym Scientific GmbH, Germany). The PCR program was carried out under thermal cycle conditions beginning with an initial preliminary denaturation at 50°C for 2 min, further 95°C for 10 min for denaturation, followed by 40 cycles of 94°C for 15 sec, 53°C for 20 sec for primer annealing and elongation at 60°C for 1 min. Additionally, all samples were analyzed by a melting temperature. In cases of forming primer dimers the reaction was repeated. A standard curve was generated by a cDNA serial dilution and amplification efficiencies were calculated to correct gene regulation. Regulation levels of each gene were calculated relative to the regulation level of the reference gene Gapdh-2 by using the comparative Ct-method (ΔΔCt-method).
Statistical analysis
Statistical analysis was performed using Prism 8 (GraphPad) software. Post-hoc comparison was applied by one-way ANOVA using Neuman-Keuls Multiple Comparison Test. Statistical significance was assumed for p<0.05 (Figure 1).
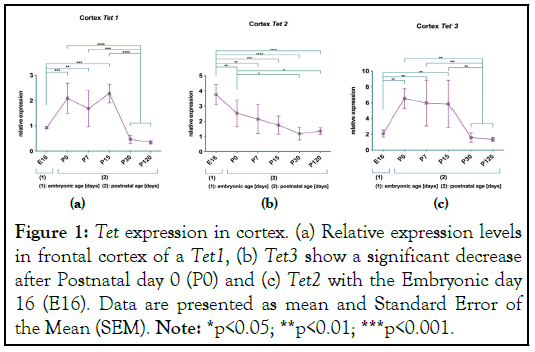
Figure 1: Tet expression in cortex. (a) Relative expression levels in frontal cortex of a Tet1, (b) Tet3 show a significant decrease after Postnatal day 0 (P0) and (c) Tet2 with the Embryonic day 16 (E16). Data are presented as mean and Standard Error of the Mean (SEM). Note: *p<0.05; **p<0.01; ***p<0.001.
The results at H3K4 show a consistent transcriptional activity of the Tet 1-3 genes over the life span. The results at H3K27 show that there is a different age dependent inactivation of Tet1 and Tet3 transcription, Tet1 gene expression in cerebellum and Tet3 gene expression in cortex are compatible with the inactivation at H3K27. The regulation of Tet2 gene show a compatibility between the Tet gene inactivations by trimethylated H3K27 and significant reduction of gene expression during aging at frontal and cerebellar cortex.
In summary hydroxylation of 5-methylcytosine during murine brain aging is regulated at H3K4 and H3K27 by Tet proteins. The regulation by trimethylated H3K4 correlates with the known increasing 5hmC amount in cortex and cerebellum (Figure 2) [8].
Figure 2: Tet expression in cerebellum. (a) Relative expression levels in cerebellum of Tet1; (b) Tet3 show a significant decrease, however with an increased peak at day P7; (c) Tet2 shows also a significant decrease in expression during aging. Data are presented as mean and Standard Error of the Mean (SEM). Note: *p<0.05; **p<0.01; ***p<0.001.
The bivalent chromatin structure is created by H3K4me3 and H3K27me3 modifications. They maintain an appropriate balance in gene regulation of pluripotent stem cells and in differentiated cells [17,18,19]. Our results confirm the functional role of H3K4me3 and H3K27me3 by generating the Ten eleven translocation (Tet) genes and 5hmC in healthy adult murine brain.
To ascertain the possibility that 5hmC is in crosstalk with the “bivalent domain”, we analyzed the ChIP-qPCR results of each Tet proteins at H3K4me3 and H3K27me3. Whole-genome analysis could detect H3K27me3 throughout the whole genome [20-22], and its methyltransferase the Polycomb Repressive Complex 2 (PRC2) to unmethylated CGIs [16,20-23]. PRC2, associated with transcriptionally silent chromatin, has histone methyltransferase activity and trimethylates histone H3 on lysine 27. Loss of PRC2 has been postulated to lose the role of gene inactivation [23]. H3K4me3 is also localized to unmethylated CGIs (Figure 3) [24].
Figure 3: Tet regulation by H3K4me3 and H3K27me3 marks.
(a) H3K27me3 correlates with inactivation status of Tet1; (b) A
significant increase of inactivation status in Tet2 is detected in
murine brain of P120; (c) Tet3 has an inverse alteration at
H3K27me3. In status of transcriptional activation (H3K4me3)
there is nearly steady level of each Tet activation without
significance. Indicated are mean and Standard Error of the
Mean (SEM). Note: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
Our data at H3K4 reveal for each Tet protein no significant effect. Given the increase of 5hmC during aging [8], we conclude that this is due to the transcriptional activity of H3K4me3.
The inactivation profile of Tet1 at H3K27me3 in our project provides the evidence that the higher or lower the induced chromatin density in aging process by H3K27me3 marks, the lower or higher the gene expression-especially at Postnatal day 120 (P120).
Regarding the results of Tet2 gene, we observe clear evidence for increase of Tet2 inactivation at H3K27me3. Our analysis of published datasets suggests that this might be the reason for reduction of Tet2 gene expression in cortex and cerebellum [11], and for an age dependent increase of 5hmC values [8]. This finding could also reveal a regulation of Tet2 by chromatin remodeling.
A fundamental issue that remains is to understand the result of regulation of Tet3 at H3K27me3. Bivalent domains were found in ES cells poising genes for activation (H3K4me3) while keeping them repressed (H3K27me3) [17]. H3K4me3 and H3K27me3 might co-occupy the same promoters with equal affinity. This occupation would establish the same results of H3K4me3 and H3K27me3 regulation. But indeed, our results in adult neuronal cells indicate that both modifications have different regulation profiles.
Basic cytology subdivides heterochromatin into constitutive and facultative heterochromatin, which becomes less-condensed euchromatin during development [25]. A comparison of the H3K27me3 modification and Tet3 gene demonstrates the association with lineage differentiation. H3K27me3 has a role in X chromosome inactivation. Gene expression of Tet3 has been observed in oocytes and cygotes [9]. Our experiments suggest that inactivation of one of the X chromosomes in female cells is the mechanism of the inactivation profile of Tet3 at H3K27me3.
The findings in understanding the regulatory function of H3K4me3 and H3K27me3 in transcription of Tet genes and in generating 5hmC. The gene regulatory role of both modifications might thus explain similar results of the next steps of 5hmC demethylation cycle (Figure 4).
Figure 4: Cytosine methylation and demethylation cycle. Cytosine(C) methylation (5mC) by DNA Methyl-Transferases (DNMT) can oxidize to 5-Hydroxymethylcytosine (5hmC) 5- carboxylcytosine (5caC) by Ten-Eleven Translocation (TET) enzymes. BER: Base Excision Repair; TDG: Thymine DNA Glycosylase.
Despite these findings some questions have to be answered. First, as Tet3 gene expression has been observed in oocytes and cygotes, experiments that separate female and male tissue will be required. Second, the binding effect of Tet proteins at antibodies needs to be focused. Regarding the detected brain regions with 5hmC, those regions are associated with neurodegenerative diseases caused by prions [26], which are misfolded proteins. Tet proteins might cause misfolding of β-sheet, as antibodies have β- sheet. The results of 5hmC may point to PrPC (cellular prion protein). Third, in addition to our different results of Tet protein regulation and expression levels, epigenetics is also the study of how behavior and environment can change gene function. Different behavioral patterns and hearing sounds which are representing by mirror neurons [27], might lead to spontaneously misfolded β-sheets (PrP prion protein) in cortex and thus might cause connection failure to grid cells. The addressing of these questions will help to understand the occurrence of brain disorders.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Yosipovitch G (2024) Regulatory Impact of Activating and Inactivating Histone Modifications for Epigenetic Control of Tet Expression during Murine Brain Development and Aging. J Epigenetics Res. 6:172
Received: 09-May-2024, Manuscript No. EROA-24-31303; Editor assigned: 10-May-2024, Pre QC No. EROA-24-31303 (PQ); Reviewed: 24-May-2024, QC No. EROA-24-31303; Revised: 31-May-2024, Manuscript No. EROA-24-31303 (R); Published: 07-Jun-2024 , DOI: 10.35248/EROA. 24.6.172
Copyright: © Yosipovitch G. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.