Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2021)Volume 14, Issue 10
Background: As we progress towards big data and large scale data analytics based genomic medicine, what should not be missed in the personalized genomic medicine context it the population specific drug prescription. Drug prescription based on population study such as demographic, gender, age, etc. is common but what can also be learnt is the drug and disease association, and that association with any subsequent follow up or non-follow up of a disease.
Methods: In the case of a non-follow up of another disease due to a certain drug-disease usage association, the drug can be a strong candidate for another disease, and thus cost effective. This can largely be possible if the fundamental biochemical pathway associated in the diseases are related or same in a key biomolecular component. Here we test the usage of drugs such as NRTIs used for HIV and Hepatitis B disease to be repositioned for usage in blindness disorders such as age related macular degeneration AMD, and find the results promising. The article also proposes new method to determine sequence of ALU isoforms in the context of AMD.
Results: The NRTIs can be safely placed for usage in combating AMD and associated eye disorders.
Conclusion: Drug Repositioning by Big Data Analytics holds great promise in cutting the time, cost and efforts in designing new molecule for combating a disease.
Macular degeneration; Repositioning drug; Interspersed signals; Bioinformatics; Big data; Large-scale data analytics
AMD: Age Related Macular Degeneration; SINES: Single Nucleotide Nuclear Interspersed Signals; NRTIs: Nucleoside Reverse Transcriptase Inhibitors; HIV: Human Immunodeficiency Virus
The initial conference publication for this work was done in the year 2018 [1]. Thereby, other scientists reported having found similar findings [2,3]. This prompted the authors to publish a follow up article in a journal, given that the discovery of NRTIs repositioning to combat AMD was first published in the article in the year 2018 [1].
Data mining such as not only genomics, but other forms of health and healthcare data can be a major factor that can be used to leverage the prescription of right drug for right patient. For instance, one can clearly see statistics of the age group that the patient is in and the likely candidate disease that are prominent for that age group, which can act as a first filter before further filtering criteria by additional symptoms can be checked in. One major challenge before treatment is the correct diagnosis itself, and that can be benefited to a large extent by generating statistical and machine learning models from the big data. Cha et al. and Brilliant et al. talk about recent methods using big data from healthcare for the methodology and benefits of drug repurposing [4,5]. If the data would be comprising of details of drug prescription history of the individuals, a time-series plot for the drug usage and follow up subsequent disease can be generated for instance at a population level to see if the usage of certain drug can likely cause another disease, and if so which ones, and so the medical practitioner can more ethically practice his profession and provide right dosage for the drug or provide a combination drug to apriori combat any possibility for further illness susceptibility. This in addition can be combined with genetic variation data of the individuals to make a more meaningful disease and drug association statistical model, many of those models will be a strong correlation anyways with a genotype given the well proven science be genes and regulation that we already know of for their missing or extra copies or a mutation. While more and more population based genome projects such as the 1000 genomes project [6] for instance are launched, and we accumulate more of genome to phenome data from the literature, it can fairly be possible to integrate the genotype phenotype databases with the clinical databases such as those maintained by hospitals, clinics and insurance agents. For example, Daffner et al. uses the PearlDiver database for a description of cost of lumbar disc herniation before surgical discectomy [7]. Lowe et al. uses another database, IBM Watson Truven Healthcare, to conduct economic modeling for certain sets of associated diseases in Urology [8]. The databases of health records by compiled insurance company suppliers such as IBM Truven and PearlDiver discussed above and many more Electronic Medical Records (EMRs) is being used for extracting knowledge in healthcare context. Developing a new drug can cost billions of dollars and more than a decade of time from the discovery phase to bringing it to the market. It is clear, that with the use of intelligent systems, and information extraction through models from the databases such as with examples stated above can be very useful in reducing time and money spent. Tools such as SasCsvToolKit (unpublished work), which can easily do several routine and complicated calculations from big data in SAS format, can be of immense use when deployed in parallel computing environment. Here we primarily focus on the scope of drug repositioning with example for AMD (age related macular degeneration) which is a leading cause of blindness among elderly people. A repositioning or repurposing of a drug can logically happen when the molecular mechanism of drug target involved in the biochemical pathway is somehow related to both the diseases, and thus this can even be numerically and statistically proven that the usage of drug for one disease can easily have an impact on the onset of other disease, as compared to those who did not take the drug. Inflammation is implicated in the progression of many diseases, and is an ideal target for therapeutic purpose. Drugs against inflammation can perhaps thus have purpose for a variety of different disorders.
ALU elements role in AMD
This article builds upon what has been submitted as a provisional patent application [9] where not only drug repositioning is talked about in the AMD context, but also a proposal to sequence and characterize the various SINES (Single Nucleotide Nuclear Interspersed Signals) (also known as ALU-elements), since one of the findings was that the reverse transcription of the SINES (ALU) elements that are present in RNA form in cytosol is perhaps a key component in AMD and other eye related disorder via formation of inflammasome [10]. ALU-elements comprise about 11% of the human genome [11]. While the quantity of cytosolic complementary DNA corresponding to ALU region can be easily determined by traditional molecular biology methods, variants of RNA-Seq in an intelligent fashion needs to be deployed to obtain the subtypes of ALU regions present in cytosol and also to differentiate it and pick the ALU sequence selectively from other RNA molecules present in the cytosol. The body of the Alu element is about 280 bases in length, formed from two diverged dimers, ancestrally derived from the 7SL RNA gene, separated by a short A-rich region [12]. As ALU sequences have a unique stretch of poly-A chains in between, which is different than polychains in the end of sequence of the RNA molecules from other sources, and having a dimer derivation, this information can be used as an intelligent means to selectively extract all the ALU molecules using magnetic beads of poly-T chain first to select the non-ALU RNA molecules and separate, and later poly-T chain with gap in between to bind the ALU RNA molecules. Let us now talk about the AMD disorder to re-focus on drug repurposing.
Age related macular degeneration
Age-related macular degeneration causes blindness and is the leading cause for it in developing nations [13-16]. AMD affects people of all ethnicities, being the main cause of vision loss as most common among Caucasians with intermediate risk in Hispanic and Asian populations and 5-fold less than Caucasians among the people of African descent [17]. Some form of AMD includes the development of abnormal blood vessels (neovascularization) leading to “wet” AMD. These blood vessels when left without treatment, result in progressive leakage due to bleeding, and irreversible scarring of the macula [18]. Wet AMD tends to occur only in 10%-15% of AMD cases, but is the more severe form as it is major cause of blindness due to AMD [19]. The wet form of AMD develop suddenly and progress to worse state even more fast, leading to vision loss [20-26]. As the population of people in the above 65 years category will rise, so will the overall burden on the nation to meet the cost associated with the treatment. Many people can be even having moderate or lesser severe AMD, but they too also need treatment due to discomfort if not blindness. There has been recent progress in the use of agents that inhibit Vascular Endothelial Growth Factor (VEGF) and has significantly helped particularly those patients who have wet AMD [27-30]. However, the treatment requires repeated injection of VEGF inhibitors directly to the eye and can be very discomfortable and expensive [31], with the cost of this treatment strategy in 2010 through Medicare part B was about USD 2 billion [32]. Herein, we test a novel hypothesis as to whether the drugs used for HIV and Hepatitis B patients such as the NRTIs (Nucleoside Reverse Transcriptase Inhibitors) can be also successfully used for patients having developed AMD and or other eye disorders. NRTIs already are used in patients with hepatitis B and human immunodeficiency virus (HIV)/ Acquired Immune Deficiency Syndrome (AIDS). This is because, it is well established that reverse transcriptase is involved in the progression of AMD, the biochemical steps that occur before the formation of inflammasome, as the NRTIs block the formation of inflammasomes [10]. It’s known that NRTIs have potential cell toxicity because they cause mitochondrial toxicity. Modified derivatives of NRTIs can be a solution if they succeed to not have that toxicity.
The purpose of this article is not to get into the details of biochemical pathway and reasoning as to why this ‘should’ happen that NRTIs can be used for combatting eye disorder or not, but given the possibility, can we deploy big data analytics for the purpose of either supporting the drug repurposing possibility in terms of numbers and statistics, or to oppose the viewpoint. It should be noted that HIV and Hepatitis B can also be treated by other means which would be then termed as Non-NRTIs or NNRTIs treatment, if the target molecule is not the reverse transcriptase. Thus, theoretically, if we can have a continuous enrollment of patient data over several years for their disease and diagnostics, then we can clearly filter out those who are either HIV or Hepatitis B infected and then record their disease progression and drug usage profile. IBM Healthcare Truven MarketScan database was chosen for healthcare data analysis. Data from the year 2006 to 2015 for a period of 10 years was used and the analysis was done on the ‘Outpatient’ category primarily. This 10 years of data is in SAS format and is of about approximately 3.5 Terabytes in size. SasCsvToolkit (unpublished, in consideration for publication) was used for data management, conversion and analysis. SasCsvToolkit has inbuilt functionalities for dealing with SAS format data for conducting various queries and operations, apart from converting the data to human readable CSV (comma separated value) format. SasCsvToolkit also helped some operations of CSV format of the data, which took about 6 Terabytes of space. Access to high performance computing system and parallel computing environment made it feasible to conduct operations on such ‘large-scale’ data systems as Simple Linux Utility for Resource Management (SLURM) was used for allocating number of computing cores, allocated memories, dependencies and compute time requirements amongst others. SLURM scripts were already incorporated in SasCsvToolkit, which needed only little adaptation for execution. International Classification of Diseases (ICD) codes revision 9 was used for all the analysis. Codes used for AMD of all types were 362.50, 362.51, 362.52, and 362.57 were used. Table 1 gives a list of all ICD-9 codes used for AMD, HIV/AIDS, and Hepatitis-B with their description. Drug codes such as national drug code numbers (NDCnum) were used for screen for drugs.
| ICD-9 Code | ICD-9 Code Description | |
|---|---|---|
| Hepatitis B | V02.61 | Hepatitis B carrier |
| 070.22 | Chronic Viral Hepatitis B with Hepatic Coma without Hepatitis Delta | |
| 070.33 | Chronic Viral Hepatitis B without Mention of Hepatic Coma with Hepatitis Delta | |
| 070.52 | Hepatitis Delta without mention of active Hepatitis B Disease or Hepatic Coma | |
| 070.31 | Viral Hepatitis B without mention of Hepatic Coma, Acute or Unspecified, with Hepatitis Delta | |
| 070.20 | Viral Hepatitis B with Hepatic Coma, Acute or Unspecified, without mention of Hepatitis Delta | |
| 070.21 | Viral Hepatitis B with Hepatic Coma, Acute or Unspecified, with Hepatitis Delta | |
| 070.23 | Chronic Viral Hepatitis B with Hepatic Coma with Hepatitis Delta | |
| 070.30 | Viral Hepatitis B without Mention of Hepatic Coma, Acute or Unspecified, without mention of Hepatitis Delta | |
| 070.32 | Chronic Viral Hepatitis B without Mention of Hepatic Coma without mention of Hepatitis Delta | |
| 070.2 | Viral Hepatitis B with Hepatic Coma | |
| 070.3 | Viral Hepatitis B without Mention of Hepatic Coma | |
| HIV/AIDS | V65.44 | Human Immunodeficiency Virus (Hiv) Counseling |
| 042 | Human Immunodeficiency Virus [Hiv] Disease | |
| V08 | Asymptomatic Human Immunodeficiency Virus [Hiv] Infection Status | |
| AMD | 362.5 | Macular Degeneration (Senile) of Retina Unspecified |
| 362.51 | Nonexudative Senile Macular Degeneration of Retina (Dry AMD) | |
| 362.52 | Exudative Senile Macular Degeneration of Retina (Wet AMD) | |
| 362.57 | Drusen (Degenerative) of Retina (Dry AMD) |
Table 1: ICD-9 codes of HIV/AIDS, Hepatitis-B and AMD used for investigation.
Broadly speaking, the steps involved screening the database for those patients having one of the ICD codes for HIV or Hepatitis B in any of the primary, secondary or subsequent diagnosis. Diagnoses of patient upto 10 values were available in the database. Unique values of the tables obtained with regard to the IDs of the patient were obtained. The patients were then filtered based on the age. From this set of data only those patients were chosen which did not have any prior history of AMD disease. Plans for check for prior history of other diseases that can be NRTI effected or interesting anyways due to inflammasome formation could also be put in place such as primary open angle glaucoma, type 2 diabetes mellitus, rheumatoid arthritis, osteoarthritis of knee, gouty arthritis, pseudogout, atherosclerosis, inflammatory bowel disease, and chronic obstructive pulmonary disease. From the group that has none of these diseases as prior history at plan of entry at the time they are diagnosed for HIV or Hepatitis B are then classified as to whether they are prescribed NRTI to combat the disease or non-NRTI. Table 2 lists some of the generic and commercial names of NRTIs and NNRTIs used for this study. From these two classification, we would then sub-classify as to how many would be the cases that develop AMD disease. These raw numbers in itself in a comparative sense from numbers in previous stage would speak about the possible use of the NRTI drug for repurposing it for AMD. However, given the results, further modeling such as multivariate logistic regression can be done for the purpose where the input variables could be those provided in the IBM Healthcare Truven MarktScan database such as age and gender, use or NRTI, etc. among others. Table 3 below broadly lists the different stages.
| NRTIs | NNRTIs | ||
|---|---|---|---|
| Generic Names | Brand Name | Generic Names | Brand Name |
| Abacavir | Ziagen | Delavirdine | Rescriptor |
| Abacavir/Dolutegravir/Lamivudine | Triumeq | Efavirenz | Sustiva |
| Abacavir/Lamivudine | Epzicom | Etravirine | Intelence |
| Abacavir/Lamivudine/Zidovudine | Trizivir | Nevirapine | Viramune |
| Adefovir | Hepsera | Rilpivirine | Edurant |
| Didanosine, DDI | Videx, Videx-EC | ||
| Emtricitabine | Emtriva | ||
| Emtricitabine/Tenofovir | Truvada, Descovy | ||
| Entecavir, ETV | Baraclude | ||
| Lamivudine, 3TC | Epivir | ||
| Lamivudine/Zidovudine | Combivir | ||
| Stavudine, d4T | Zerit | ||
| Tenofovir | Viread | ||
| Zalcitabine | Hivid | ||
| Zidovudine, azidothymidine (AZT) | Retrovir |
Table 2: Drug names used for NRTIs and NNRTIs for analysis.
Though the stages of selection looks straightforward, the amount of coding needed to be done for different ICD-9 codes, and prior history as well as subsequent development checks, given that the raw file names need to be also preserved in order to ensure there is no ambiguity and artifact in solutions generated needed, would be laborious for a big data system in SAS format, and the SasCsvToolkit comes with handy set of features for easy implementation of the work in a high performance computing system environment. It can well be possible to rename the input files and create scripts that can do things in an automated way, but in any case the scripts have to be created at first place and then there are chances of results getting mixed up due to changed names of input files. The SasCsvToolkit comes with tools where you can use the file names as it is without modifications and submit feature via SLURM script is already also provided.
The results generated using the workflow until the linear filtering which is stage 4, which has been also double checked for correctness is shown in Table 4, for only HIV patient datasets. It is by no surprise that the drop in number is way too high in the initial stages than at later stages. Thereafter, the results of both outpatient and facility header categories were pooled in for stage 5 analysis to see how many of them ever used an NRTI drug to deal with HIV infection. The results are shown in Figure 1. As expected, a high proportion of people were prescribed with NRTI drug. The next level comprises of taking each of the two categories of patients and looking for any subsequent disease condition for AMD. Figure 2 shows it for those who took NRTI as drug and Figure 3 shows it for those who did not take the NRTI drug but took some other form of medication. In both the population set, people getting AMD at a later date as per the limitation in the amount of data years we had, only 10 years, was less likely. However, even with the limited data of 10 years, we could clearly see that while the percentage of people developing AMD was only about 0.9% for those who took NRTI as drug, the percentage was fairly high relatively speaking, about 17%, for those who took other forms of drug but not NRTI.
| Stage | Data |
|---|---|
| Stage 1 | All Patients |
| Stage 2 | HIV and Hepatitis B infected |
| Stage 3 | Filtered for those which are more than 50 years of age and have had AMD |
| Stage 4 | Filter for those which have had no prior AMD disorder at the time of HIV or Hepatitis B diagnosis for first time |
| Stage 5 | Divide the dataset into groups that were prescribed NRTIs and those that were not prescribed NRTIs but possibly something else |
| Stage 6 | Divide each group into number of cases of subsequent development of disease or not (altogether 4 categories will be obtained) |
Table 3: Big data work in healthcare context here for NRTI drug repositioning for AMD patients.
| Results of HIV outpatient and facility header files from IBM Watson Truven MarketScan workflow | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Outpatient | |||||||||||
| Stage | Year | ||||||||||
| 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | ||
| Raw data | 23700516 | 26309515 | 36687933 | 40307778 | 39039010 | 42511346 | 43707058 | 34847335 | 36429764 | 22319004 | |
| HIV relevant ICD9 | 33847 | 38690 | 60211 | 63919 | 61380 | 68512 | 72737 | 62045 | 66908 | 40620 | |
| Age>34 | 26924 | 31221 | 48624 | 52235 | 50178 | 55155 | 58397 | 49802 | 53000 | 31952 | |
| Age>49 | 8489 | 10480 | 17414 | 19722 | 19785 | 23232 | 26127 | 24012 | 26620 | 16372 | |
| After removal of entries with prior AMD history | 8463 | 10401 | 17251 | 19545 | 19549 | 22938 | 25776 | 23649 | 26175 | 16089 | |
| Facility header | |||||||||||
| Stage | Year | ||||||||||
| 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | ||
| Raw data | 10672036 | 11866867 | 1.7E+07 | 18983537 | 18192497 | 19866463 | 20153228 | 15718520 | 16627720 | 10253584 | |
| HIV relevant ICD9 | 11562 | 13699 | 22251 | 24898 | 24473 | 27488 | 27984 | 23232 | 26846 | 15897 | |
| Age>34 | 9357 | 11207 | 18196 | 20458 | 20210 | 22391 | 22687 | 18618 | 21338 | 12540 | |
| Age>49 | 3113 | 3976 | 6876 | 8145 | 8332 | 9799 | 10594 | 9419 | 11154 | 6713 | |
| After removal of entries with prior AMD history | 3099 | 3939 | 6806 | 8069 | 8243 | 9665 | 10436 | 9261 | 10961 | 6594 | |
Table 4: Filtering the IBM Watson Truven MarketScan database in outpatient and facility header criteria, by various screening conditions as listed in method section, for HIV patients for year 2006-2015.
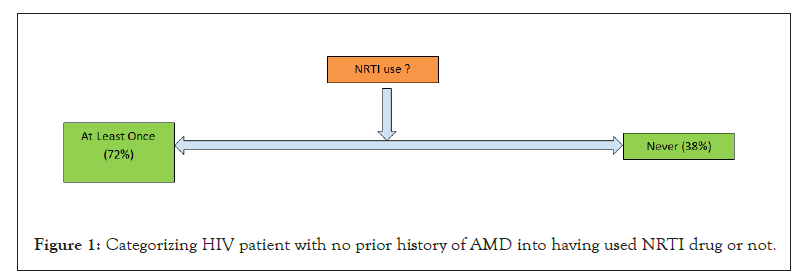
Figure 1: Categorizing HIV patient with no prior history of AMD into having used NRTI drug or not.
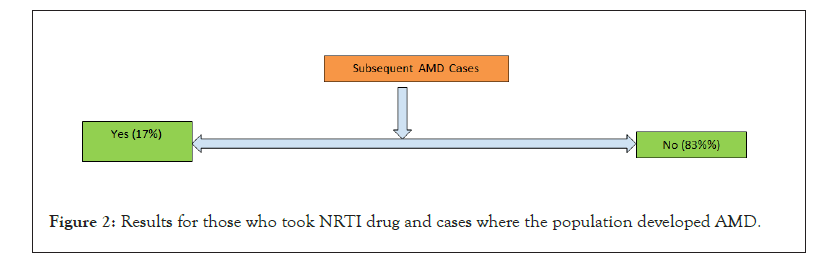
Figure 2: Results for those who took NRTI drug and cases where the population developed AMD.
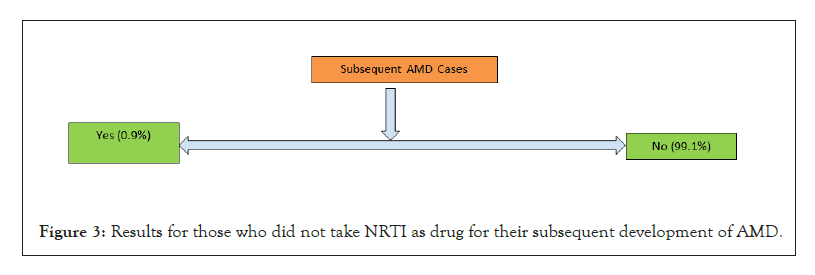
Figure 3: Results for those who did not take NRTI as drug for their subsequent development of AMD.
At this point it should be also noted that the continuity of the data and patient record would be skewed, as only the people diagnosed in 2006 will have a maximum of 10 years of follow up diagnosis, and records of people diagnosed in 2015 will have zero years of follow up. The patients with records between 2006 and 2015 will have a value in between 0 and 10 years as maximum continuity, and thus, in cases where the disease actually shows up after say 10 years, they would easily get missed out by the current dataset we purchased. As we were limited by the budget of not having able to purchase more years of data, our results thus got limited by what we could show as evidence for this limited set of data. Furthermore, even if we manage to get more data such as from other data vendors, it is also more important that the patient Id number remains more or less the same in these 10 plus years to trace his medical conditions throughout, and sadly, as people either change their job several times within 10 years, or at least their healthcare insurance providers, which are the suppliers to the data vendors, even though one might buy several terabytes of data, eventually the continuity over several years, makes the effective data available for analysis such as this very limited.
We have demonstrated that big data analytics can be used very systematically and schematically with the help of domain knowledge such as in biochemical pathway in medical context, to show wonders. The better the data volume and the required kind of data, which in our case is the year-over-year continuity, the better and more reliable results will be. The results for Table 4 were double checked, and the results generated for Figures 1, 2, and 3 were generated using standard approach using inbuilt functions in SasCsvToolkit, and will require in future double confirmation by using more data sources. More yearly continuity of data sources needs to be obtained. Analysis of this kind needs to be first also done over Hepatitis B patients who have similar reverse transcriptase biochemical mechanism of action, and should also be done for many more eye diseases such as glaucoma. Future work also can be done in taking the input column variables and see how the output results of getting AMD is illustrated in each of the derived datasets such as by multilogistic regression. The study was limited in the sense that the data was primarily for US market, however, given the diversity of population in US, it can fairly represent the broad spectrum of population. Future work would involve conducting something similar in other geographies. The SasCsvToolkit would also be released soon for public usage, as soon as the licensing setup work for the tool is completed.
A modified non-toxic form of NRTI can be used as a drug to combat AMD and related disorders. This work having done purely by big data analytics, saves several years of drug discovery process and encourages more application of methods for drug repositioning by data science techniques.
Consent to publication was taken at the time when bio-samples were being collected.
All supporting data are provided in this article.
None to be declared.
None
All work, idea generation, coding, analysis of results and writing the paper has been done by the first author.
Citation: Singh AN (2021) Repurposing Anti-Inflammasome NRTIs for AMD by Data Mining. J Proteomics Bioinform. 14: 555.
Received: 22-Sep-2021 Accepted: 06-Oct-2021 Published: 13-Oct-2021 , DOI: 10.35248/0974-276X.21.14.555
Copyright: © 2021 Singh AN. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.