Enzyme Engineering
Open Access
ISSN: 2329-6674
ISSN: 2329-6674
Short Communication - (2022)Volume 11, Issue 4
SARS-CoV2 main protease is important for viral replication and one of the most potential targets for drug development in this current pandemic. Drug repurposing is a promising field to provide potential short-term acceptable therapy for management of coronavirus till a specific anti-viral for coronavirus is developed. In-silico drug repurposing screening is the current fastest way to repurpose drugs by targeting active sites in fraction of seconds. In this study, SARS-CoV2 main protease is being targeted by 1050 FDA-approved drugs to inhibit its activity thereby interfering with viral replication. Chemotherapeutic drugs and anti-retroviral drugs have shown potential binding as inhibitor. In-vitro and clinical trials required to establish final fact.
SARS-CoV2; Drug repurposing; FDA; Chemotherapeutic; Anti-retroviral
SARS-CoV2 main protease is important for viral replication and one of the most potential targets for drug development in this current pandemic. Discovery of new drugs takes years of development and trials. The current pandemic situation makes it more dicult to arrange resources and conduct studies. Drug repurposing is a promising field to provide potential short-term acceptable therapy for management of coronavirus till a specific anti-viral for coronavirus is developed. In-silico drug repurposing screening is the current fastest way to repurpose drugs by targeting active sites in fraction of seconds. In this study, SARS-CoV2 main protease is being targeted by 1050 FDA-approved drugs to inhibit its activity thereby interfering with viral replication. Choosing FDA-approved drugs for this study is important, so that any potential result obtained can be procured easily without delay. The procedure used in this study had been defined in a previous study [1] and necessary permission was obtained to reproduce it here. (See Procedure in the Supplementary Files) The three-dimensional structure of SARS-CoV2 main protease with inhibitor was downloaded from Protein Data Bank (PDB id: 6WTT) (Figure 1) [2,3]. For molecular docking, three-dimensional structures of 841 compounds were obtained from Zinc15 database [4]. The compounds constituted only those which were approved by FDA. This step was taken so as to facilitate easy availability after clinical trials. 6WTT structure constitutes three chains A, B and C. In order to obtain the active site of the receptor, Drug Discovery Studio software [5] was utilized to find the interactions between the inhibitor and amino acids. These amino acids would serve as the desired target in molecular docking. PyRx software which includes within itself Autodock Vina and Open Babel was used for molecular docking [6-8]. Since all three chains and their corresponding ligands were similar, only one chain was used as the target. The chain was extracted from the entire structure and it was cleared of all ligands and water molecules. The cleaned structure was converted into an Autodock macromolecule by addition of Hydrogen atoms and partial charges. The ligand compounds from Zinc15 database were brought to the minimum energy configuration and were converted in Auto dock ligands by using Open Babel. The vina search space parameters were set to the following values so as to include all targets amino acids.
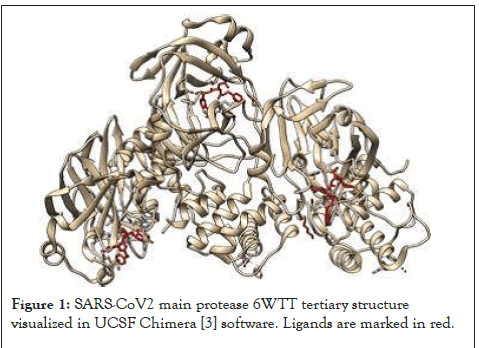
Figure 1: SARS-CoV2 main protease 6WTT tertiary structure visualized in UCSF Chimera [3] software. Ligands are marked in red.
Center_x=2.92116253491
Center_y=26.9617112837
Center_z=-10.4416644449
Size_x=18.1979693434
Size_y=19.9112571778
Size_z=28.9662108454
Exhaustiveness was set to 4 for faster computation. All the above operations were performed on a Windows 10 64-bit operating system. The entire docking took about 8 hours with 100% CPU utilization (Figure 2). The results of docking obtained were sorted on basis of binding affinity. Lower the binding afinity, more stringer is the interaction. For every ligand configuration, only those which had root mean square deviation (RMSD) value 0.0 were chosen. The interactions of receptor with every ligand were visualized using Drug Discovery Studio. The number of desired target amino acids interacted was noted and final result was obtained. If any ligand showed unfavorable interactions, it was excluded to improve accuracy of the study. The pocket site of the inhibitor was visualized and the following amino acids were found to constitute the active site. Standard amino acid abbreviations are used.
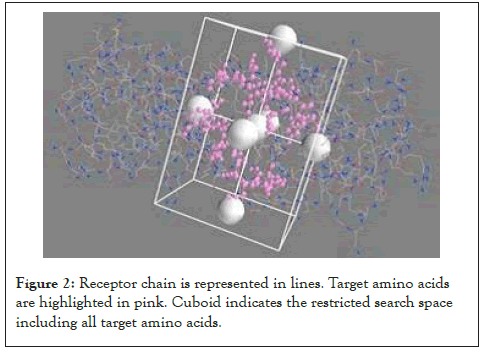
Figure 2: Receptor chain is represented in lines. Target amino acids are highlighted in pink. Cuboid indicates the restricted search space including all target amino acids.
Numbers indicate their position:
1. Pro 168
2. Gln 189
Page 4/17
3. Met 49
4. Phe 140
5. His 41
6. His 172
7. Glu 166
8. His 164
The binding afinity of top 20 of 841 ligands docked with receptor chain are arranged in the below Table 1 along with number of target amino acids it is bound to out of 8 mentioned above. It is first sorted on the basis of decreasing order of interactions, then on the basis of increasing order of binding afinity up to -8.1 kcal/mol. The current therapeutic use has also been mentioned. Their interactions are depicted below in Figures 3-5 with amino acids of receptor chain (circles with sequence position of that amino acid. For all the above interactions, green bonds are the conventional hydrogen bonds. Pi-alkyl bonds are depicted in pink. Pi-sulphur bonds are depicted in yellow. Pi-donor hydrogen bond is depicted in light blue. Pi-Pi T shaped bonds are depicted in magenta. Halogen bond is depicted in blue. Carbon-Hydrogen bonds are depicted in light blue. Pisigma bond is depicted in purple. Literature review of the obtained drugs was conducted to establish any known studies proving its eficacy against coronavirus. Celsentri, saquinavir, nilotinib and risperdal had been reported to even inhibit SARS-CoV2 entry by targeting the receptor-binding domain [1]. Celsentri, lurasidone, paliperidone, argatroban, azelastine, dihydroergotamine, conivaptan, rolapitant and saquinavir have also been reported in other in-silico studies to inhibit protease [9-11,12-17]. Nilotinib has shown to inhibit MERS-CoV at 5.5 micromolar and SARSCoV at 2.1 micromolar concentration [11]. Candesartan has been theoretically studied for treatment of SARS-CoV2 [12]. However, there were no relevant studies found for rest of the drugs linking them to coronavirus.
| Ligand | Binding affinity | Name | Use | Active site out of 8 |
|---|---|---|---|---|
| chain_A_ZINC000003817234_u ff_E=983.51 | -8.5 | Celsentri | HIV | 5 |
| chain_A_ZINC000003914596_u ff_E=525.60 | -9.1 | Saquinavir | HIV | 4 |
| chain_A_ZINC000006716957_u ff_E=651.98 | -8.3 | Nilotinib | chemotherapy | 4 |
| chain_A_ZINC000003816292_u ff_E=1638.28 | -8.1 | Lenvatinib | chemotherapy | 4 |
| chain_A_ZINC000003827556_u ff_E=1074.02 | -8.1 | Delafloxacin | anti-biotic | 4 |
| chain_A_ZINC000004074875_u ff_E=1089.05 | -8.1 | Candesartan | Hypertensi on | 4 |
| chain_A_ZINC000003816514_u ff_E=531.67 | -9 | Rolapitant | anti- emetic | 3 |
| chain_A_ZINC000003978005_uff_E=940.58 | -8.7 | Dihydroergot amine | Migraine | 3 |
| chain_A_ZINC000012503187_u ff_E=1067.62 | -8.6 | Conivaptan | Hyponatre mia | 3 |
| chain_A_ZINC000003927822_u ff_E=1108.64 | -8.5 | Lurasidone | mental disorder | 3 |
| chain_A_ZINC000004214700_u ff_E=566.16 | -8.5 | Palperidone | mental disorder | 3 |
| chain_A_ZINC000000538312_u ff_E=544.51 | -8.3 | Risperdal | mental disorder | 3 |
| chain_A_ZINC000003920266_u ff_E=480.19 | -8.3 | Idarubicin | Chemothe rapy | 3 |
| chain_A_ZINC000014210642_u ff_E=1228.10 | -8.3 | Edarbi | Hypertensi on | 3 |
| chain_A_ZINC000000601229_u ff_E=407.32 | -8.2 | Azelastine | anti- histaminic | 3 |
| chain_A_ZINC000000537805_u ff_E=659.74 | -8.1 | Diabeta | type 2 diabetes | 3 |
| chain_A_ZINC000003938684_u ff_E=968.87 | -9.8 | Toposar | Chemothe rapy | 2 |
| chain_A_ZINC000004175630_u ff_E=542.57 | -8.5 | Orap | Tourette Synd | 2 |
| chain_A_ZINC000003917708_u ff_E=550.40 | -8.3 | Daunorubicin | Chemothe rapy | 2 |
| chain_A_ZINC000003807172_u ff_E=822.43 | -8.1 | Argatroban | Anticoagulant | 2 |
Table 1: Top 20 ligands with name according to Zinc15 database arranged according to number of active site interactions and biding affinity.
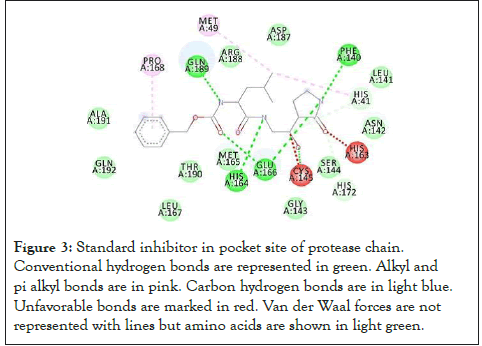
Figure 3: Standard inhibitor in pocket site of protease chain. Conventional hydrogen bonds are represented in green. Alkyl and pi alkyl bonds are in pink. Carbon hydrogen bonds are in light blue. Unfavorable bonds are marked in red. Van der Waal forces are not represented with lines but amino acids are shown in light green.

Figure 4: Standard inhibitor in pocket site of receptor represented by hydrophobicity surface.
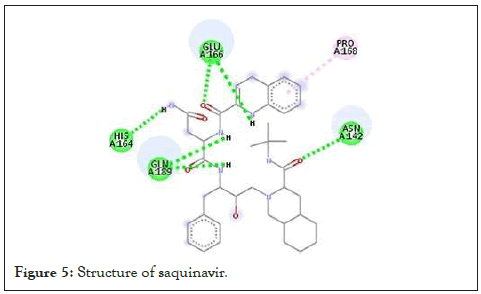
Figure 5: Structure of saquinavir.
These drugs must be pushed to in-vitro screening and clinical trials as treatment options or chemoprophylaxis. In this study I have screened a large library of FDA-approved drugs and put forth promising results. At times of this ongoing pandemic, this study might contribute in the fight against coronavirus. The results though encouraging, must be tested in laboratory before proceeding with human trials and public use because of the limitations of in-silico approach.
Source of funding
Nil.
Ethical approval
Not required.
Conflict of interest
The author hereby declares no conflict of interests.
[Crossref]
[Crossref]
[Crossref]
Citation: Sharp K (2022) Repurposing Inhibitors for SARS-CoV2 Main Protease Enz Eng. 11:189.
Received: 01-Jun-2022, Manuscript No. EEG-22-17694; Editor assigned: 03-Jun-2022, Pre QC No. EEG-22-17694 (PQ); Reviewed: 21-Jun-2022, QC No. EEG-22-17694; Revised: 28-Jun-2022, Manuscript No. EEG-22-17694 (R); Published: 05-Jul-2022 , DOI: 10.35841/2165-8056.22.11.189
Copyright: © 2022 Sharp K. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.