Journal of Tumor Research
Open Access
ISSN: 2684-1258
ISSN: 2684-1258
Research Article - (2024)Volume 10, Issue 3
Craniopharyngioma although benign, is one of the most difficult tumors to resect and manage. The fluid inside the tumors is known to accumulate even after aspiration leading to tumor recurrence. Spontaneous rupture of the tumor leads to leakage of the cyst fluid to the surrounding tissue and is known to cause aseptic meningitis. This study aimed to identify toxic proteins in the cyst fluid which could be responsible for altering the normal physiology of the surrounding brain tissue. We first characterized the proteomic content of the cyst fluid collected from 12 patients by tandem mass spectrometry which revealed 91 common proteins. These proteins were then subjected to bioinformatics analysis to identify the biological processes, cell components and molecular functions of the proteins and were further compared with the proteins involved in neurodegenerative diseases to narrow our search for toxic proteins. 46 out of the 91 proteins were taken for functional enrichment on Cytoscape. Functional enrichment then revealed a total of 21 toxic proteins in the fluid which could play a role in the regulation of cell death or are involved in cell killing. The interaction of a repertoire of proteins involved in metastasis, inflammation, extracellular matrix reorganization and cell death as evidenced by the protein-protein interaction network suggests the interplay of the proteins in the cyst fluid, which could directly or indirectly be cytotoxic. Data are available via proteome exchange with identifier PXD051226.
Craniopharyngioma; Cyst Fluid; Proteomics; Gene Ontology; Protein-Protein Interaction Network
CP: Craniopharyngioma; ACP: Adamantinomatous Craniopharyngioma; PCP: Papillary Craniopharyngioma; CTNNB1: Catenin beta-1; MAPK: Mitogen-Activated Protein Kinase; CSF: Cerebrospinal Fluid; LC-MS: Liquid Chromatography-tandem Mass Spectrometry; MS: Mass Spectrometry; GO: Gene Ontology; PTM: Post-Translational Modifications
Craniopharyngiomas (CPs) are rare benign non-neuroepithelial neoplasms arising from the remnants of Rathke’s pouch. CPs constitute 1.2%-4.6% of all intracranial tumors and account for 0.5-2.5 cases per million population annually [1]. CPs have a bimodal age distribution, with one peak in ages 5-15 years and another peak in ages 45-60 years [1,2]. CPs are generally found in the sellar or suprasellar regions of the brain, although cases of ectopic CPs have also been reported [3]. CPs can arise from anywhere in the craniopharyngeal duct and can extend up to the third ventricle. Anatomically, CPs are very close to some of the brain’s critical regions, including the hypothalamus, optic chiasm, cranial nerves, the third ventricle and major blood vessels. Primary symptoms include headache and nausea caused by increased intracranial pressure [4]. The pressure of the tumor mass on the optic chiasm leads to reduced visual acuity and visual field defects [5]. The proximity of the tumor to these vital structures makes the management of CPs a challenging task.
The 5th edition of the World Health Organization (WHO) classification of Central Nervous System Tumors (CNS5, 2021), classifies CPs into two types- Adamantinomatous Craniopharyngioma (ACP) and Papillary Craniopharyngioma (PCP). ACPs are characterized by Catenin Beta-1 (CTNNB1) mutations in exon 3 encoding the beta-catenin gene, which increases beta-catenin stability and leads to the activation of the Wnt pathway [6,7]. About 95% of PCPs are characterized by V-Raf Murine Sarcoma Viral Oncogene Homolog B (BRAF) Valine600Glutamine (V600E) mutations [8]. This mutation is known to activate the Mitogen Activated Protein Kinase (MAPK) pathway and results in cell proliferation and tumor growth. ACPs have both a solid and a cystic component [9]. The solid component of ACPs consists of epithelial cell nests embedded in loose connective tissue. The degeneration of the stellate cells surrounding these epithelial nests leads to the formation of micro cysts in the tumor [10]. The cyst fluid resembles a ‘motor-oil-like’ dense fluid composed of bone residues, calcium flecks and high concentrations of lipids, especially cholesterol [11,12]. The fluid is brownish-yellow in color due to its high cholesterol content. Microscopic evaluation reveals the presence of keratin, ghost cells and dystrophic calcifications [12].
Studies to characterize the proteome of cystic fluid are still underway and have been enriched by the reports of a few labs in the last decade. One of the first contributions to the analysis of cystic fluid revealed the presence of beta-2 microglobulin in the fluid as well as in the Cerebrospinal Fluid (CSF) of CP patients [13]. Arefyeva et al., reported elevated lactate levels in the fluid compared to blood serum, which was in accordance with the pH of the fluid, which was found to be lower than that of the serum [14]. Mori et al., hypothesized the role of Interleukin 6 (IL-6) in inflammatory reactions on the border of tumor tissue and healthy brain tissue [15]. Suck et al., studied the biochemical composition of the cyst fluid, that revealed elevated levels of calcium, cholesterol, glucose, low and high-density lipids and transaminases [16].
In vitro studies have demonstrated the toxic effects of cyst fluid on hypothalamic neurons and glial cells. Ghosh et al., reported a decline in the viability of primary hypothalamic cultures from neonatal rat brains due to cyst fluid exposure. The same study also reported morphological changes in the plasma membrane of the cells in the form of membrane blebbing suggesting the initiation of apoptosis [17]. Cyst fluid has also been suggested to cause neuro inflammation through increased production of IL-6 [18]. The same study also reported enhanced interaction of Cluster of Differentiation 74 (CD74) with Amyloid precursor protein (APP) between the inflammatory-activated microglia and hypothalamic neurons. Production of several inflammatory markers including Glial Fibrillary Acidic Protein (GFAP), endothelial Nitric Oxide Synthase (e-NOS) and Nuclear Factor Kappa B (NF-kB) was stimulated by cyst fluid exposure, in addition to its involvement in inducing oxidative stress in the cortical region of the rat brain [19].
The gross-total resection of CPs remains a challenging task even today and as a sub-total resection leads to tumor recurrence, it is empirically necessary to study the protein content of the cystic fluid to develop intra-cystic therapies that could provide much-needed relief to the patients suffering from this disease worldwide. In this study, we analyzed the complete proteomic profile of cystic fluid from 12 patients suffering from CP using LC-MS/MS and assessed it using Gene Ontology (GO) enrichment analysis. The goal of this study was to search for proteins in the CP cyst fluid that could potentially be cytotoxic to their surrounding tissues.
Materials
Bicinchoninic Acid Assay (BCA) protein estimation kit was from Thermo Scientific™ Pierce™ BCA Protein Assay kits, C18 stage-tips were prepared using 3M™ Empore™ C18 disks. Vacuum concentrator was from Thermo Scientific™ Savant™ SPD1010 SpeedVac™ and sequencing grade trypsin was from Promega™ San Luis Obispo, CA, USA. C18 trap column used was Acclaim PepMap™ 100 with dimensions of 75 µm × 2 cm, nano viper 2 Pack, particle size 3 µm and pore size 100 Å from Thermo Scientific™. Analytical column used was PepMap™ Rapid Separation Liquid Chromatography (RSLC C18) with dimensions of 75 µm × 50 cm, particle size 2 µm and pore size 100 Å from Thermo Scientific™. Dithiothreitol, iodoacetamide, acetone, formic acid and all other chemicals were purchased from Sigma.
Clinical samples
Cyst fluid was collected from patients suffering from CP, at the National Institute of Mental Health and Neurosciences (NIMHANS), Bangalore after acquiring their consent and approval from the Institutional Ethics Committee. 12 patients were recruited for the study, out of which 4 were females. The patient details pertaining to age, sex, symptoms experienced at the time of diagnosis, location of the tumor and histopathology are mentioned in (Table 1). All the experiments with human subjects were performed in accordance with the Declaration of Helsinki. Cyst fluid samples collected from the patients were spun at 10,000 rpm for 5 minutes to remove the blood contaminants and cellular debris. The supernatant was collected and snap-frozen at -80°C until further use.
| Patient no. | Age (years) | Sex | Symptoms | Location of the tumor | Histopathology |
|---|---|---|---|---|---|
| P1 | 7 | M | Headache, visual deterioration | Sellar, suprasellar | Adamantinomatous craniopharyngioma |
| P2 | 13 | M | Headache, vomiting and decreased vision in left eye |
Sellar, suprasellar | Adamantinomatous craniopharyngioma |
| P3 | 12 | M | Headache, vomiting | Suprasellar | Adamantinomatous craniopharyngioma |
| P4 | 10 | F | Headache, progressive diminution of vision in both eyes | Suprasellar | Adamantinomatous craniopharyngioma |
| P5 | 9 | M | Headache, vomiting | Suprasellar | Adamantinomatous craniopharyngioma |
| P6 | 3 | F | Headache, vomiting | Suprasellar | Adamantinomatous craniopharyngioma |
| P7 | 4 | M | Headache, vomiting | Multicompartment-sella, Suprasellar, temporal, posterior fossa | Adamantinomatous craniopharyngioma |
| P8 | 17 | M | Painless vision loss in both eyes | Suprasellar | Papillary craniopharyngioma |
| P9 | 7 | M | Headache, vomiting | Suprasellar | Adamantinomatous craniopharyngioma |
| P10 | 23 | M | Headache, vomiting, diabetes Insipidus | Sellar, suprasellar | Adamantinomatous craniopharyngioma |
| P11 | 69 | F | Vision loss in both eyes | Sellar, suprasellar | Adamantinomatous craniopharyngioma |
| P12 | 9 | F | Headache, vomiting | Sellar, suprasellar | Adamantinomatous craniopharyngioma |
Note: M: Male; F: Female.
Table 1: Details of patients recruited in the study.
Sample preparation
About 50 µL of the cyst fluid was taken and lysed in 2% Sodium Dodecyl Sulfate (SDS) buffer (2% SDS w/v in 50 mM Triethylammonium Bicarbonate (TEABC)) for the extraction of the proteins. Protein estimation was done using the BCA protein estimation kit. Equal amounts of proteins from each sample were taken for reduction and alkylation using 5 mM Dithiothreitol and 10 mM iodoacetamide, respectively. The proteins were precipitated using ice-cold acetone. Sequencing grade trypsin was used at a ratio of 1:20 following overnight incubation at 37°C to digest the proteins. The resulting peptides were desalted using C18 stage-tips and dried using a vacuum concentrator.
LC-MS/MS analysis
The dried peptides were reconstituted in 0.1% formic acid before injecting them into the mass spectrometer. Data were acquired on Thermo Scientific™- Orbitrap Fusion™ Tribrid™ MS coupled with Thermo Scientific™ Easy n-LC 1000 nano-flow liquid chromatography system. The peptides were loaded onto a C18 trap column (Acclaim PepMap™ 100 Å, 75 µm × 2 cm, nano viper 2 Pk, C18, 3 µm, 100 Å) at a flow rate of 6 µL/min and separated on an analytical column (PepMap™ RSLC C18, 75 µm × 50 cm, 2 µm, 100 Å) with a gradient 8-35% of 100% acetonitrile in 0.1% formic acid over 104 minutes at a flow rate of 280 nL/min.
LC-MS/MS analysis was done on an Orbitrap analyzer in data-dependent mode with a resolution of 120,000 at 200 m/z for full scan and 30,000 at 200 m/z for MS/MS. High-energy Collision Dissociation (HCD) was used with Collision Energy (CE) 34% for fragmentation of the precursor ions selected at full scan. Dynamic exclusion of 30 seconds was applied for the fragmented ions. Automated Gain Control (AGC) for full scan was set to 4e5 and for MS2 it was set to 1e5 with a maximum time of 50 ms and 54 ms respectively.
Data analysis
Data from LC-MS/MS analysis were searched using Proteome Discoverer (version 2.1) using National Center for Biotechnology Information (NCBI) RefSeq database (release 89). We used sequestHT as the search engine. The search parameters included oxidation on methionine; acetylation on the N-terminal of protein as dynamic modification; carbamidomethylation on cysteine as static modification and trypsin as the enzyme with a maximum of two missed cleavages. Precursor mass tolerance of 10 ppm and a fragment mass tolerance of ± 0.05 Da were applied. False Discovery Rate (FDR) of 1% was applied at the Peptide Spectrum Match (PSM), peptide level and protein level.
The mass spectrometry proteomics data has been deposited to the Proteome Exchange Consortium via the PRIDE partner repository with the dataset identifier PXD051226.
Statistics and bioinformatics analysis
Gene ontology of the proteomics data from mass spectrometry was then carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID) gene ontology tool available online. An online tool called “Interacti Venn” was used to generate the Venn diagram for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. In addition, ToppGene analysis of the proteins was performed to understand the correlation of the proteins with neurodegenerative diseases. The list of diseases given by ToppGene had an FDR B and H correction cutoff <0.05. A Protein-Protein Interaction (PPI) network was generated using the Search Tool for the Retrieval of Interacting Genes/Proteins database for the proteins found to be involved in neurodegenerative diseases. The parameters for generating the string network on the STRING database were kept as high confidence (0.700) for interaction score and network edges set to confidence. This network was then exported to Cytoscape (version 3.9.1) to visualize the string network. The functional enrichment performed on Cytoscape gave us the proteins enriched for biological processes with an FDR value <0.05.
Analysis of mass spectrometric data
To study the proteomic profile of the cyst fluid, we subjected the samples to LC-MS/MS. The MS data was then used to further study the GO enrichment analysis using DAVID for KEGG pathways, biological processes, cell components, molecular functions and Post-Translational Modifications (PTMs).
ESI-Orbitrap-LC-MS/MS was used to analyze for a complete proteome screening for all the samples and the generated data was used for further study. The Mass Spectrometric (MS) data of the fluid samples were filtered for good spectral quality with high confidence and supported with at least two unique peptides and Peptide Spectral Match (PSMs) respectively. Table 2 shows the number of filtered proteins from each sample. A total of 91 common proteins (after excluding duplicates) were identified among all the samples (Figure 1).
| Patient No. | Number of hits from LC-MS/MS data | Filtered proteins |
|---|---|---|
| P1 | 205 | 153 |
| P2 | 214 | 151 |
| P3 | 235 | 158 |
| P4 | 179 | 133 |
| P5 | 222 | 156 |
| P6 | 236 | 174 |
| P7 | 430 | 199 |
| P8 | 210 | 148 |
| P9 | 234 | 167 |
| P10 | 227 | 164 |
| P11 | 212 | 158 |
| P12 | 211 | 147 |
Note: LC-MS: Liquid Chromatography-tandem Mass Spectrometry; MS: Mass Spectrometry
Table 2: Number of hits and filtered proteins for each sample.
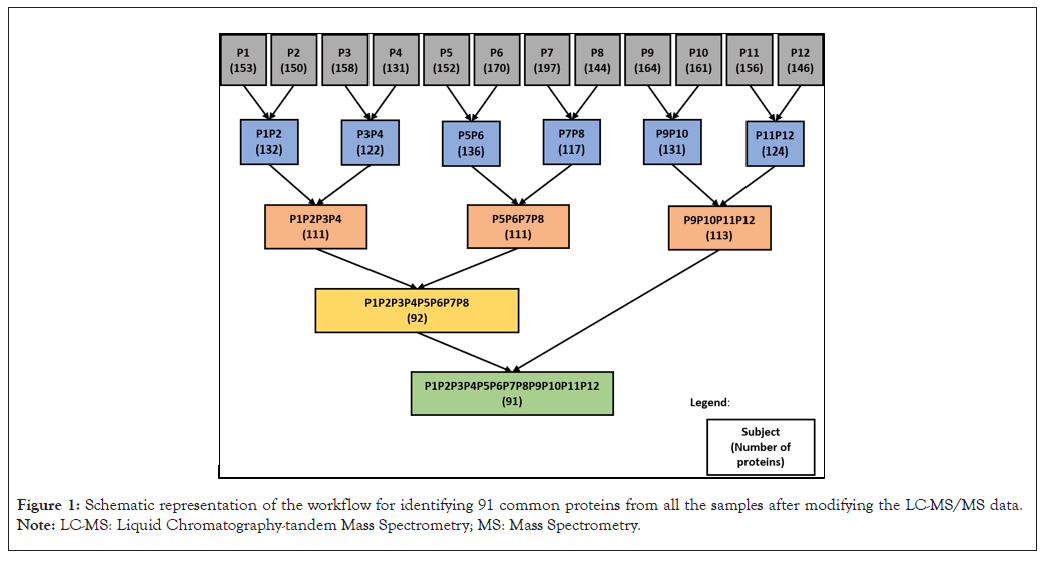
Figure 1: Schematic representation of the workflow for identifying 91 common proteins from all the samples after modifying the LC-MS/MS data. Note: LC-MS: Liquid Chromatography-tandem Mass Spectrometry; MS: Mass Spectrometry.
Kyoto encyclopedia of genes and genomes analysis for proteins involved in specific pathways
KEGG pathway analysis was performed to investigate the specific pathways implicated in CP. The 91 common proteins were subjected to KEGG pathway analysis on DAVID. 61/91 (67.0%) of the proteins were found to be significantly enriched for the KEGG pathway analysis. The output showed us that 37 proteins were involved in various inflammatory pathways and 14 proteins were involved in various metabolic pathways. 4 proteins were found to be common in both the inflammatory and metabolic pathways. Venn diagram (Figure 2) shows the number of proteins involved in inflammatory pathways and metabolic pathways. Details of these proteins are mentioned in Table 3. The inflammatory pathways included complement and coagulation cascades, neutrophil extracellular trap formation, phagosome and platelet activation. The metabolic pathways included cholesterol metabolism, Extracellular Matrix (ECM)-receptor interaction, focal adhesion and regulation of the actin cytoskeleton.
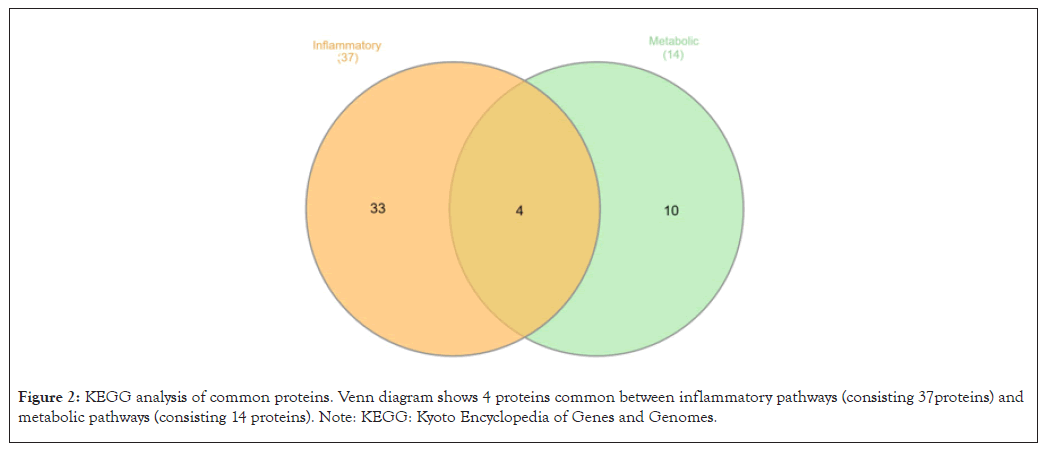
Figure 2: KEGG analysis of common proteins. Venn diagram shows 4 proteins common between inflammatory pathways (consisting 37 proteins) and metabolic pathways (consisting 14 proteins). Note: KEGG: Kyoto Encyclopedia of Genes and Genomes.
| Gene symbol | Descriptions | Inflammatory pathways | Metabolic pathways |
|---|---|---|---|
| A2M | Alpha-2-macroglobulin isoform x1 | P | A |
| ACTG1 | Actin, cytoplasmic 2 | P | P |
| APOA1 | Apolipoprotein a-i isoform 1 preproprotein | A | P |
| APOA2 | Apolipoprotein a-ii preproprotein | A | P |
| APOB | Apolipoprotein b-100 precursor | A | P |
| APOE | Apolipoprotein e isoform a precursor | A | P |
| APOH | Beta-2-glycoprotein 1 precursor | A | P |
| C1QB | Complement c1q subcomponent subunit b isoform x1 | P | A |
| C1QC | Complement c1q subcomponent subunit c isoform 1 precursor | P | A |
| C1R | Complement c1r subcomponent isoform 2 preproprotein | P | A |
| C2 | Complement c2 isoform 1 preproprotein | P | A |
| C3 | Complement c3 preproprotein | P | A |
| C4B | Complement c4-b preproprotein | P | A |
| C4BPA | C4b-binding protein alpha chain isoform x1 | P | A |
| C5 | Complement c5 isoform 2 | P | A |
| C6 | Complement component c6 isoform x1 | P | A |
| C7 | Complement component c7 precursor | P | A |
| C8A | Complement component c8 alpha chain preproprotein | P | A |
| C8B | Complement component c8 beta chain isoform x1 | P | A |
| C8G | Complement component c8 gamma chain precursor | P | A |
| C9 | Complement component c9 preproprotein | P | A |
| CD14 | Monocyte differentiation antigen cd14 precursor | P | A |
| CFB | Complement factor b preproprotein | P | A |
| CFH | Complement factor h isoform a precursor | P | A |
| CFHR1 | Complement factor h-related protein 1 precursor | P | A |
| CFI | Complement factor i isoform 1 preproprotein | P | A |
| CLU | Clusterin isoform x1 | P | O |
| F2 | Prothrombin isoform 1 preproprotein | P | P |
| FGA | Fibrinogen alpha chain isoform alpha-e preproprotein | P | A |
| FGB | Fibrinogen beta chain isoform 1 preproprotein | P | A |
| FGG | Fibrinogen gamma chain isoform gamma-b precursor | P | A |
| FN1 | Fibronectin isoform 1 precursor | A | P |
| GSN | Gelsolin isoform a precursor | A | P |
| KLKB1 | Plasma kallikrein isoform x1 | P | A |
| KNG1 | Kininogen-1 isoform 2 precursor | P | P |
| LAMA3 | Laminin subunit alpha-3 isoform x1 | A | P |
| LAMB3 | Laminin subunit beta-3 isoform x1 | A | P |
| LOC102723407 | Immunoglobulin heavy variable 4-38-2-like | P | A |
| PLG | Plasminogen isoform 1 precursor | P | A |
| PLTP | Phospholipid transfer protein isoform a precursor | A | P |
| PROS1 | Vitamin K-dependent protein S isoform 1 precursor | P | A |
| SERPINA1 | Alpha-1-antitrypsin precursor | P | A |
| SERPINC1 | Antithrombin-III isoform 1 precursor | P | A |
| SERPIND1 | Heparin cofactor 2 precursor | P | A |
| SERPINF2 | Alpha-2-antiplasmin isoform X2 | P | A |
| SERPING1 | Plasma protease C1 inhibitor precursor | P | A |
| VTN | Vitronectin precursor | P | P |
Note: P: Present; A: Absent
Table 3: List of proteins found in inflammatory and metabolic pathways.
Gene ontology analysis for biological processes, cellular components, molecular functions and post-translational modifications
The common proteins were analyzed for their enriched Biological Processes (BPs), Cellular Components (CCs), Molecular Functions (MFs) and Post-Translational Modifications (PTMs). These GO terms give us a rough idea of the protein’s involvement in various cellular and molecular functions spatially. The BPs and MFs together help us determine the activities of the gene product that are relevant to the functioning of the cell.
A total of 89/91 proteins (97.8%) were found to be significantly enriched across the BPs. The proteins in the BPs were classified under those involved in signaling, regulatory or response, protein homeostasis, transport, cell cycle, development and differentiation, lysis or disassembly and others (Figure 3A). 90/91 (98.9%) proteins were found to be significantly enriched for CCs. Many proteins were classified under extracellular proteins followed by membrane proteins, cell organelle proteins and others (Figure 3B). These results are in accordance with the fluid which is an extracellular component of the tumor and contains a high concentration of lipids. 87/91 (95.6%) proteins were found to be enriched for MFs. The proteins under MFs were categorized as activity, receptor binding, binding and others (Figure 3C). The proteins were further analyzed for their PTMs which revealed most proteins as glycoproteins followed by proteins with disulfide bonds, pyrrolidone carboxylic acid, cleavage on a pair of basic residues, sulfation, hydroxylation, oxidation, etc. (Figure 3D). A total of 90/91 (98.9%) proteins were found to be significantly enriched for PTMs.
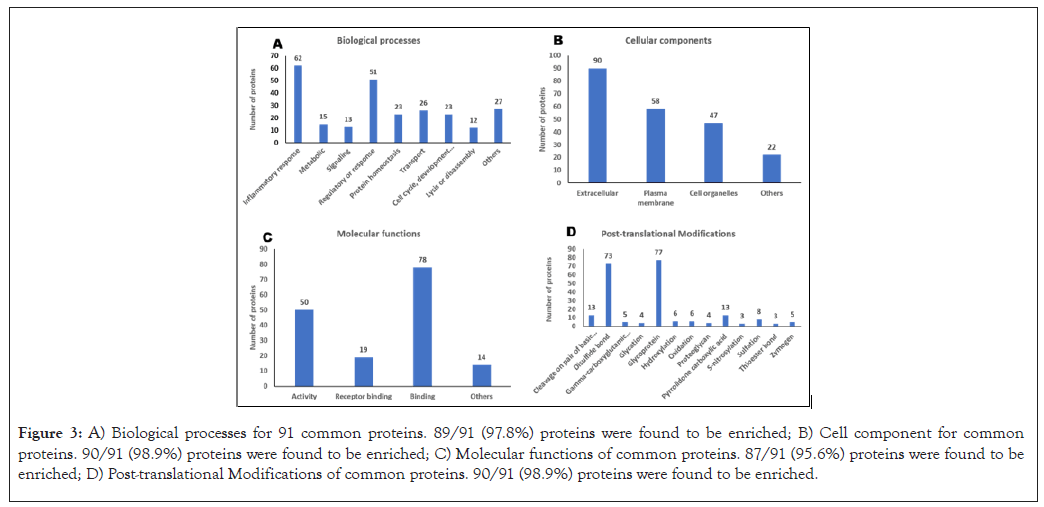
Figure 3: A) Biological processes for 91 common proteins. 89/91 (97.8%) proteins were found to be enriched; B) Cell component for common proteins. 90/91 (98.9%) proteins were found to be enriched; C) Molecular functions of common proteins. 87/91 (95.6%) proteins were found to be enriched; D) Post-translational Modifications of common proteins. 90/91 (98.9%) proteins were found to be enriched.
ToppGene and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis for identifying potentially toxic proteins
To narrow down our search for potentially toxic proteins in the cyst fluid, we compared the 91 common proteins with proteins implicated to be involved in various neurodegenerative diseases by subjecting them to ToppGene analysis. Multiple Sclerosis; Presenile Dementia; Alzheimer’s Disease, Late-Onset; Parkinson's Disease; Amyloid Neuropathies; Cerebral Amyloid Angiopathy; Dementia; Amyotrophic Lateral Sclerosis; and Senile Plaques were some of the neurodegenerative diseases characterized by ToppGene analysis with p-values <1.95E-08. 46 of the 91 proteins were found to be involved in the aforementioned neurodegenerative diseases.
The 91 common proteins were then subjected to STRING analysis to generate a PPI network with 91 nodes and 459 edges and PPI enrichment p-value <1.0E-16. This PPI network was then exported to Cytoscape to visualize the protein-protein interaction. The PPI network (Figure 4) highlights the 46 proteins (identified in neurodegenerative diseases by the ToppGene analysis) from the 91 common proteins enriched by the STRING analysis. The thickness of the edges in the network is directly proportional to the interaction score, while the direction of the interaction is indicated by the arrow of the edges. These 46 proteins were then used to generate a PPI network with 46 nodes and 451 edges, PPI enrichment p-value <1.0E-16 on the STRING database. Functional enrichment was performed on the network after exporting it to Cytoscape. Functional enrichment revealed 21 proteins involved in cell death/cell killing or the regulation of cell death (Figure 5). Table 4 lists the details of these 21 proteins.
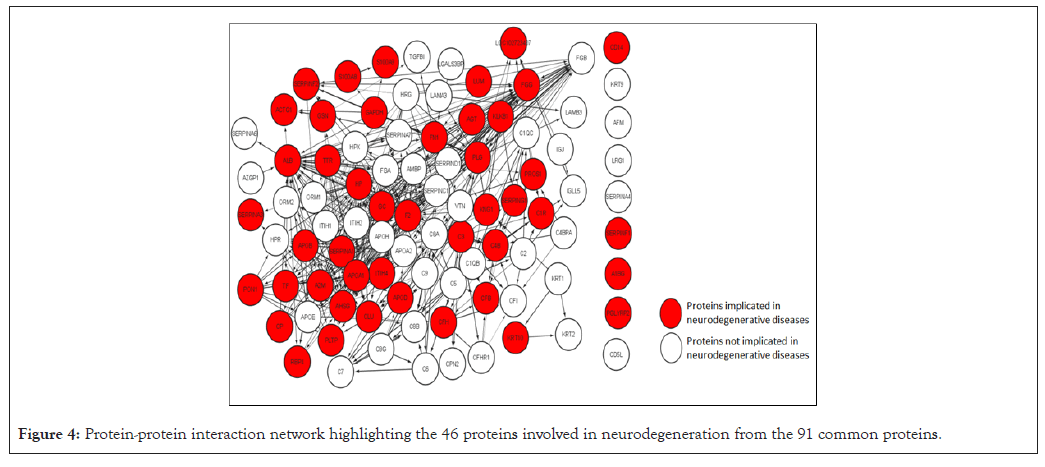
Figure 4: Protein-protein interaction network highlighting the 46 proteins involved in neurodegeneration from the 91 common proteins.
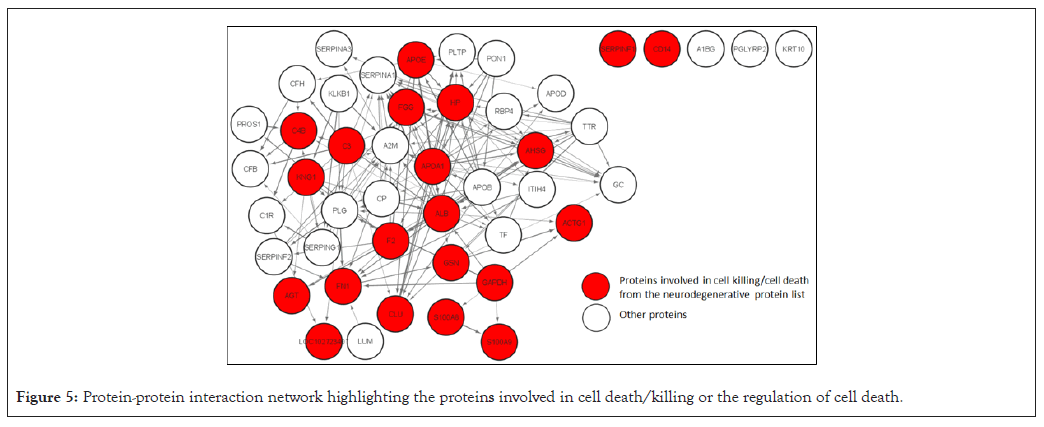
Figure 5: Protein-protein interaction network highlighting the proteins involved in cell death/killing or the regulation of cell death.
| Gene symbol | Description |
|---|---|
| Actg1 | Actin, cytoplasmic 2 |
| Agt | Angiotensinogen preproprotein |
| Ahsg | Alpha-2-hs-glycoprotein isoform 2 preproprotein |
| Alb | Serum albumin preproprotein |
| Apoa1 | Apolipoprotein a-i isoform 1 preproprotein |
| Apoe | Apolipoprotein e isoform a precursor |
| C3 | Complement c3 preproprotein |
| C4b | Complement c4-b preproprotein |
| Cd14 | Monocyte differentiation antigen cd14 precursor |
| Clu | Clusterin isoform x1 |
| F2 | Prothrombin isoform 1 preproprotein |
| Fgg | Fibrinogen gamma chain isoform gamma-b precursor |
| Fn1 | Fibronectin isoform 1 precursor |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase isoform 1 |
| Gsn | Gelsolin isoform a precursor |
| Hp | Haptoglobin isoform 1 preproprotein |
| Kng1 | Kininogen-1 isoform 2 precursor |
| Loc102723407 | Immunoglobulin heavy variable 4-38-2-like |
| S100a8 | Protein s100-a8 isoform d |
| S100a9 | Protein s100-a9 |
| Serpinf1 | Pigment epithelium-derived factor isoform 1 precursor |
Table 4: List of proteins involved in cell death/cell killing or regulation of cell death.
Spillage of cystic fluid during surgeries or their spontaneous rupture is implicated to cause adverse effects on the brain tissue from cellular necrosis to impairment of neurocognitive functioning. The complete protein content of the cyst fluid has yet to be characterized even though major work in the field has been going on for decades. A study by Donson et al., identified elevated levels of several inflammatory markers like Interleukin (IL) IL-6, IL-8, CXC motif chemokine ligand 1(CXCL1) and IL-10 in both ACP cyst fluid and solid tumors [20]. Another study identified the activation of the inflammasome in ACP cyst fluid and solid tumor. This study reported the overexpression of inflammatory genes like IL-1B, IL-18, IL-6, IL-8 and IL-10 [21]. Intra-cystic therapies and radiotherapies have been widely used to avoid post-surgical complications like loss of anterior pituitary hormonal function, diabetes insipidus, life-long morbid obesity and neurocognitive changes. Radio therapeutic agents (90 Yttrium and 32 phosphorus) are used intra-cystically but have been reported to cause neuronal toxicity and brain damage or even death [22]. Intra-cystic therapeutic agent, bleomycin was the first drug to be used widely in chemotherapies, but now it has been replaced with interferon-alpha due to the former’s neurotoxic effects. However, recent literature has also pointed to the potential neurotoxic effects of interferon alpha in patients with prior radiation exposure [23].
Our mass spectrometric data revealed the presence of Alpha-2-HS-Glycoprotein Isoform 2 Preprotein (AHSG), Apolipoprotein A-I Isoform 1 Preprotein (APOA-I), Apolipoprotein A-II Preprotein (APOA-II) and Clusterin (CLU) which is in accordance to that reported by Martelli et al., [24]. The functional enrichment analysis performed on Cytoscape categorized AHSG, APOA-I, Apolipoprotein E (APOE), CLU, Complement C3 preproprotein (C3), Actin, Cytoplasmic 2 (ACTG1), Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH), protein S100-A8 isoform d (S100A8) and protein S100-A9 (S100A9) into biological processes related to the regulation of cell death. Serine Proteinase Inhibitor A3 (SERPINA3), a 47.6 kDa protein, is usually expressed in the liver but in certain cases, it is found to be expressed in astrocytes, muscle cells and monocytes [25]. It is found to be involved in the complement cascade, inflammation and wound healing [26]. GO enrichment analysis suggests that SERPINA3 negatively regulates the endopeptidase activity and is involved in the inflammatory response in addition to protein binding functions. SERPINA3 is a biomarker for progressive multiple sclerosis and is upregulated in Alzheimer’s disease [27,28]. SERPINA3 upregulation is associated with glioma progression as it contributes to the invasive behavior of glioblastoma cells by proteolytically degrading the extracellular matrix [26]. SERPINA3 is also found to be upregulated in colon cancer [29], cholangiocarcinoma tissues and endometrial cancers [30,31]. AHSG is a 39.3 kDa glycoprotein with 2 polypeptide chains linked with a disulfide bond. It is suggested to be involved in bone metabolism regulation, vascular calcification, insulin resistance and protease activity [32]. In our results, AHSG was categorized as an extracellular protein in cellular component with involvement in the inflammatory response and positive regulation of phagocytosis under biological processes. ASHG is thought to be over secreted due to the accumulation of lipids in adipose tissue. AHSG is suggested to drive tumor progression in pancreatic tumors and prostate cancers, while it is believed to be involved in angiogenesis in glioblastomas. Synthesis of AHSG by tumor cells has been reported to cause tumor growth and promote metastasis [33].
APOE is a 39 kDa protein that transports cholesterol, essential for the maintenance of myelin and neuronal membranes in the central and peripheral nervous systems [34]. GO enrichment analysis revealed that APOE is involved in the efflux of cholesterol from the cells in addition to lipid and receptor binding activities, which could explain the high lipid content of the cyst fluid. Functional enrichment also found APOE to be involved in the positive regulation of cell death and regulation of the apoptotic process. APOE is essential for cell proliferation and survival in ovarian carcinoma [35]. APOE is said to be interfering with beta-amyloid clearance by altering the enzyme which degrades beta-amyloid, thereby resulting in plaque formation in Alzheimer’s. Further, it was shown to enhance oligomeric beta-amyloid toxicity by directing it to synapses [36]. CLU is a secreted hetero-dimeric glycoprotein of 75-80 kDa, with multi-faceted functions including lipid transport and cell adhesion, while in its truncated form of 55 kDa, it is targeted to the nucleus where it functions as a death signal [37]. GO enrichment analysis revealed the role of clusterin in complement activation and innate immune response. Functional enrichment found CLU to be involved in the regulation of the apoptotic signaling pathway, regulation of cell death and regulation of the apoptotic process. High glial expression of CLU in ALS patients with 43 kDa Transactive response Deoxyribonucleic Acid Binding Protein (TDP-43) pathology and cognitive deficits causes axonal degeneration and cell death leading to reactive gliosis [38]. CLU is expressed in neurons and astroglia and is found to be elevated in Alzheimer’s disease, where it co-localizes with amyloid-beta. Intra-cellular accumulation of CLU results in binding with amyloid-beta thereby, preventing amyloid-beta proteolysis and leading to cytotoxic effects [39]. Overexpression of CLU results in tumorigenesis in ovarian and prostate cancer, while it leads to cell proliferation, migration and metastasis in renal cell carcinoma [40-42].
C3, a 187 kDa protein that plays a key role in complement activation, is found to be upregulated in microglia of Alzheimer’s and was found to be elevated in ALS patients [43,44]. C3 is believed to be involved in cellular proliferation and sustaining angiogenesis and oncogenesis and it is reported to be used as a marker for early pancreatic cancer [45]. GO enrichment analysis found C3 to be involved in negatively regulating endopeptidase activity, in addition to being a key molecule involved in inflammatory pathways, while functional enrichment found C3 to be involved in the positive regulation of phagocytosis and cell killing. Overexpression of C3 is suggested to activate the Janus kinase (JAK)/ Signal Transducer and Activator of Transcription 3 (STAT3) pathway with downstream effects leading to cell proliferation, apoptosis, angiogenesis, tumor invasion and metastasis [46, 47]. ACTG1 is a 41.7 kDa cytosolic protein that functions in non-muscle cells. ACTG1 was found to be involved in the inflammatory response and cell cycle, development and differentiation in addition to being a regulatory or response protein. Functional enrichment found ACTG1 to be involved in phagocytosis. Gamma-actin is implicated to be involved in cell migration in neuroblastoma cells [48]. Overexpression of ACTG1 is thought to inhibit the mitochondrial apoptotic pathway, upregulate cyclin-dependent kinases, thereby resulting in the proliferation of cells and promoting the Warburg effect, thus leading to tumorigenesis [49].
In addition to these aforementioned proteins, we also found some other proteins with interesting functions. GAPDH, a homotetramer of 36 kDa, once thought to be a “housekeeping gene”, has multiple functions like cytoskeletal dynamics, vesicle transport and regulation of cell survival and cell death [50,51]. GO enrichment analysis found GAPDH to be involved in the inflammatory response, regulatory or response functions and protein homeostasis, among others. Functional enrichment found GAPDH to be involved in cell killing and regulation of cell death. Continuous oxidative stress has been reported to cause conformational changes in GAPDH, thereby aiding its aggregation and eventually promoting cell death [52]. Exogenous GAPDH has been reported to induce loss of inner transmembrane potential leading to the release of cytochrome c and apoptosis-inducing factor through the outer membrane thus leading to apoptosis in cancer cells [53]. Several studies have reported the correlation between GAPDH expression and formation and malignancy of lung cancer [54,55]. S100A9 is a calcium-binding protein and may exist as a homodimer or a heterodimer with an S100A8 partner, which interestingly was also found in our mass spectrometric analysis. Both S100A8 and S100A9 were found to be involved in the inflammatory response, signaling and protein homeostasis, among others in the GO enrichment analysis. S100A8 and S100A9 were found to be involved in most of the cell death/cell killing functions vise, positive regulation of cell death, positive regulation of apoptotic signaling pathway, regulation of cell death and positive regulation of intrinsic apoptotic signaling pathway. S100A9 has been reported to function with the Tumor Necrosis Factor-alpha (TNF- α) and Transforming Growth Factor-beta (TGF-β) pathway that promotes cell growth and metastasis in non-small cell lung adenocarcinoma and breast cancer [56,57]. S100A8 and S100A9 levels were found to be increased in hepatocellular carcinoma and S100A9 was reported to activate reactive oxidative species pathway thereby inhibiting apoptosis [58]. Secretion of S100A8- S100A9 from cortical neurons have dual roles of recruiting myeloid cells in the cortex leading to inflammation and interacting with the receptor of advanced glycation end product to promote astrocyte growth eventually leading to gliosis [59].
CP is a challenging tumor to resect and manage, with the cystic fluid accumulating inside even after aspiration, leading to tumor recurrence. Unconstrained burst of the tumor can cause aseptic meningitis due to spilling of the cyst fluid on the surrounding tissue. This study aimed to identify toxic proteins in the cyst fluid, which could alter the normal physiology of the surrounding brain tissue. The proteomic content of cyst fluid collected from 12 patients was characterized by tandem mass spectrometry, revealing 91 common proteins. Bioinformatics analysis identified biological processes, cell components and molecular functions of these proteins, which were compared with proteins involved in neurodegenerative diseases. Functional enrichment on Cytoscape revealed 21 toxic proteins in the fluid, potentially playing a role in cell death regulation or cell killing. The interaction of proteins involved in metastasis, inflammation, extracellular matrix reorganization and cell death suggests the potential for cytotoxic interactions in cyst fluid.
Regarding the possible limitations of the study, as 11 of the 12 subjects had ACP, this study does not consider the histopathological differences that could arise in the samples. This study also does not take into account the differences that could arise due to gender. As the source of cyst fluid is a tumor tissue, the lack of ideal control in the study does not allow us to assess the expression levels of the proteins in the sample.
It is evident from the proteomic profile that the cystic component of the tumor is a dynamic system and plays an essential role in tumor growth and progression. Most of the proteins in the cyst fluid were found to be involved in lipid metabolism, inflammatory response, proteolysis, calcium ion binding and protein binding which is in accordance with the cyst fluid content. The proteins present in the fluid not only have roles in various inflammatory and metabolic pathways which are severely altered in tumor physiology, but are also cytotoxic to neurons and microglia as observed from our bioinformatics analysis. Our mass spectrometric data has unveiled a few proteins which might be the reason for the neurotoxicity of the cyst fluid. As bioinformatics analysis was the primary investigating tool for accessing the cytotoxicity of the fluid content, studies to understand the functions and the exact roles of proteins implicated in tumor microenvironment are needed.
All the samples were obtained after taking consent from the patients. All human experiments were performed in accordance with the Declaration of Helsinki.
Manish Beniwal and Bhupesh Mehta acknowledge NIMHANS proteomics facility.
Bhupesh Mehta, Manish Beniwal, KVL Narasingha Rao, Yogananda S. Markandeya and Periyasamy Govindaraj contributed to the conceptualization of the study, experimental design and manuscript revision. MB and KVLN contributed to sample collection from the subjects. Omkar Shirke, Vivek Ghose and Rahul R. Patil contributed in performing experiments, data analysis and manuscript writing.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Narasingha Rao KVL, Shirke O, Ghose V, Patil RR, Markandeya YS, Govindaraj P, et al. (2024). Revealing Toxic Proteins of the Craniopharyngioma Cyst Fluid: A Proteomics Approach. J Tumor Res. 10:234.
Received: 02-Sep-2024, Manuscript No. JTDR-24-31753; Editor assigned: 05-Sep-2024, Pre QC No. JTDR-24-31753 (PQ); Reviewed: 18-Sep-2024, QC No. JTDR-24-31753; Revised: 25-Sep-2024, Manuscript No. JTDR-24-31753 (R); Published: 04-Oct-2024 , DOI: 10.35248/2684-1258.24.10.234
Copyright: © 2024 Narasingha Rao KVL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.