Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research - (2024)Volume 17, Issue 1
Objective: To identify of vital genes in the blood exosome related to the process of gastric carcinogenesis and help to reduce mortality rates through early diagnosis and the development of new anticancer therapies.
Method: The Ribo-Nucleic Acids (RNAs) data of blood exosomes from patients with Gastric Cancer (GC) and healthy controls were downlinked from exoRBase database and the differential expression of messenger RNA (mRNA), long non coding RNA (lncRNA) and circular RNA (circRNA) were analyzed using R language. Then the relevant RNAs and their corresponding micro RNA (miRNA) data predicted by Encyclopedia of RNA Interactomes (ENCORI), miRcode and other databases, were imported into the competing endogenous RNA (ceRNA) network. Finally, The Database for Annotation, Visualization and Integrated Discovery (DAVID) was accessed to investigate the Differentially Expressed mRNA (DEmRNAs), Gene Ontology (GO) annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Results: A total of 62 mRNAs, 3 lncRNAs and 15 circRNAs were differentially expressed. The ceRNA network was constructed with cytoscape software, including 192 mRNA nodes, 32 lncRNA nodes, 28 circRNA nodes and 152 miRNA nodes, with the top 10 hub genes EMSY, Zinc finger E-Box-Binding homeobox 2 (ZEB 2), Ligand Dependent Nuclear Receptor Corepressor (LCOR), Major Facilitator Superfamily Domain containing 14A (MFSD 14A), ERBB2 Interacting Protein (ERBB2IP), hsa-miR-363-3p, hsa-miR-137, hsa-miR-27a-3p, hsa-miR-23b-3p and hsa_circ_0000038. According to GO annotation, the biological processes mainly in cytoplasmic translation, the cell component were mostly in the ribosome and its subunit and the major molecular functions included structural constituent of ribosome, etc. The KEGG revealed that the DEmRNA were primarily enriched in mRNA monitoring pathway.
Conclusion: A ceRNA network in the blood exosome of GC were built, the hub genes were screened out, the biological process, cell component, molecular function and enrichment pathway of DEmRNA were explored, which proved that the ribosome biogenesis was a major player in the exosome of gastric cancer and this provide precise targets for making a diagnosis and giving treatment of GC.
Gastric cancer; Exosome; Enrichment analysis; Ribosome biogenesis
Gastric Cancer (GC) is a kind of cancer that occurs in the stomach, mostly from gastric mucosal epithelial cells. The most common pathological type is adenocarcinoma. The long-term survival rate (>5 years) of early GC after surgery can reach 90.9%-100% [1-3]. However, there is still no effective treatment for advanced GC. Even with various treatments, the survival rate of 5-year is still less than 30%. The incidence rate of GC ranks the fifth in malignant tumors and the third in mortality worldwide. Early detection of early GC or postoperative recurrence of GC is still the focus of cancer prevention and treatment [4].
The exosome is extracellular vesicle with the diameter of 30- 150 nm, which is released into the extracellular matrix after the outer membrane fuses with the cell membrane [5]. They have lipid bilayer and contain mRNA, lncRNA, circRNA and transfer RNA (tRNA), etc., which are vital for tumor cell biology, such as proliferation and metastasis, also can serve as prognostic markers and/or grading basis for patients with tumors, belong to the research field of liquid biopsy [6-12].
In this study, we obtained the RNA (including mRNA, lncRNA and circRNA) information of plasma exosome samples from patients with GC and healthy people from exoRBase database, screened the differentially exosomal RNAs in GC using bioinformatics methods, constructed a network related to GC with the competitive endogenous RNA (ceRNA) [13-15]. The functional enrichment analysis of mRNA is also carried out in order to discover more new GC markers and provide guidance for early detection, early identification and early therapy of GC.
Data source
The expression profiles of RNA in GC and healthy blood samples were download from exoRBase 2.0. The data deadline was December 27, 2022. The data format was pretreated. 9 blood samples from GC patients and 118 healthy blood samples were obtained, involving mRNA and lncRNA 35517 and circRNAs 79084.
Filtering of Diversely Emanated Genes (DEGs)
The R language "limma" package was applied to take the mean value and the "sva" package was employed for batch correction to identify the DEGs. The filtering condition for DEGs was p-value<0.05. The corresponding volcano map was drawn with the "heat map" package of top 20 genes with obvious differences.
Prediction of miRNAs and construction of ceRNA networks
Target scan human and miRanda in ENCORI (https://starbase.sysu.edu.cn/) were employed to combinedly forecast the differentially express mRNAs-bound miRNAs. The miRNA target function in ENCORI was applied to forecast the miRNA binding to DEcircRNAs and the lncRNAs binding miRNAs were predicted in miRcode (http://www.mircode.org/). Finally, the prediction data of miRNAs and the corresponding mRNAs, circRNAs and lncRNAs were input into the cytoscape software (version 3.7.2) to display graphically the ceRNA network.
Hub gene sifting
The cytoHubba plugin of cytoscape software was utilized to pick the top 10 nodes out as hub gene by the betweenness algorithm and the topology index of the betweenness of each hub node was used to plot in excel.
Functional enrichment unpacking
The DEmRNAs were input to DAVID database (https://david.ncifcrf.gov/), with the identifier as "OFHCIAL-GENE-SYMBOL" and the species as "Homo sapiens", GO annotations and KEGG pathway analysis were carried according to the conditions of P<0.05 and False Discovery Rate (FDR)<0.05 (FDR is the p-value after correction and calculation).
Statistical methods
All statistical analyses were completed in R4.2.2 statistical software. The mean ± standard deviation was employed to display the measurement data and t-test or analysis of variance was utilized to perform the statistical test. P<0.05 means the difference was statistically significant.
Results of DEGs screening
62 DemRNAs and 3 differentially expressed lncRNAs 2 (all up-regulated genes) were screened out, with the Heterogeneous Nuclear Ribo-Nucleo Protein K (HNRNPK) as the most significant mRNA, AC011450.1 exhibiting the most relevant lncRNA. The DEGs heat map is as documented (Figure 1). One down regulated circRNAs (hsa_circ_0007476, genomic position: chr4:53414615-53428183, genomic length: 13568, spliced length: 356) with differential expression (DEcircRNAs) as shown (Figure 2).
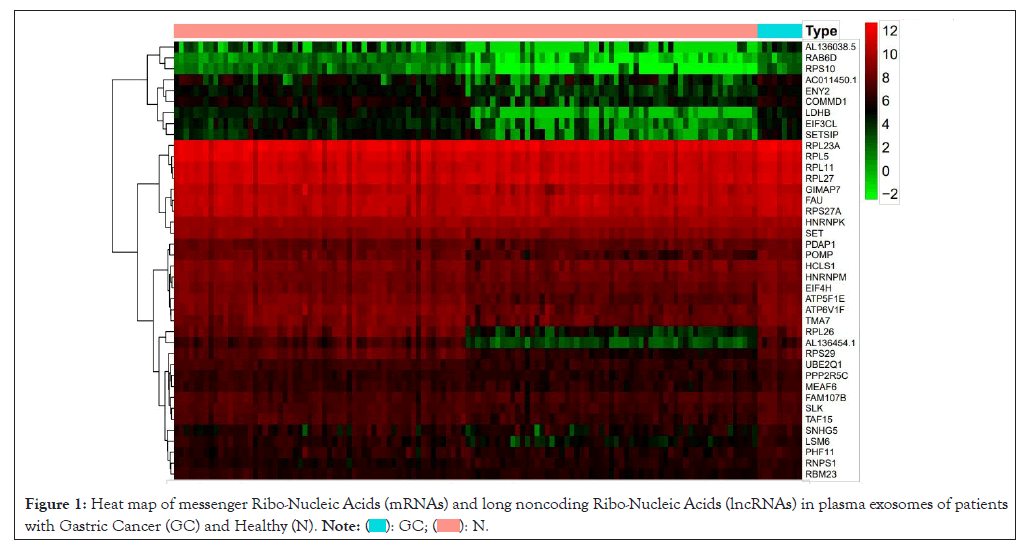
Figure 1: Heat map of messenger Ribo-Nucleic Acids (mRNAs) and long noncoding Ribo-Nucleic Acids (lncRNAs) in plasma exosomes of patients
with Gastric Cancer (GC) and Healthy (N). 
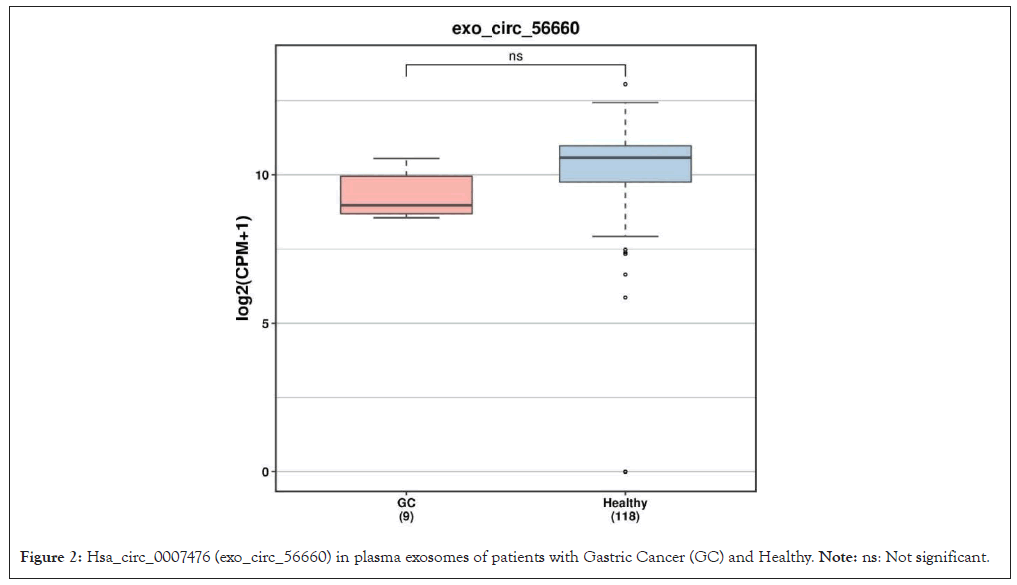
Figure 2: Hsa_circ_0007476 (exo_circ_56660) in plasma exosomes of patients with Gastric Cancer (GC) and Healthy. Note: ns: Not significant.
Establish of exosomal ceRNA network
According to the data of DEmRNAs, DElncRNAs, DEcircRNAs and their binding miRNAs, a competitiveness endogenous RNA network was constructed, with 192 mRNA nodes (involving 22 mRNA), 32 lncRNA nodes (1 lncRNA involved), 28 circRNA nodes (involving 1 circRNA) and 151 miRNA nodes (Figure 3). The top 10 pivot genes with the highest betweenness scores were distinguished by cytoscape software, including 5 mRNAs, 4 miRNAs and 1 circRNA (Figure 4).
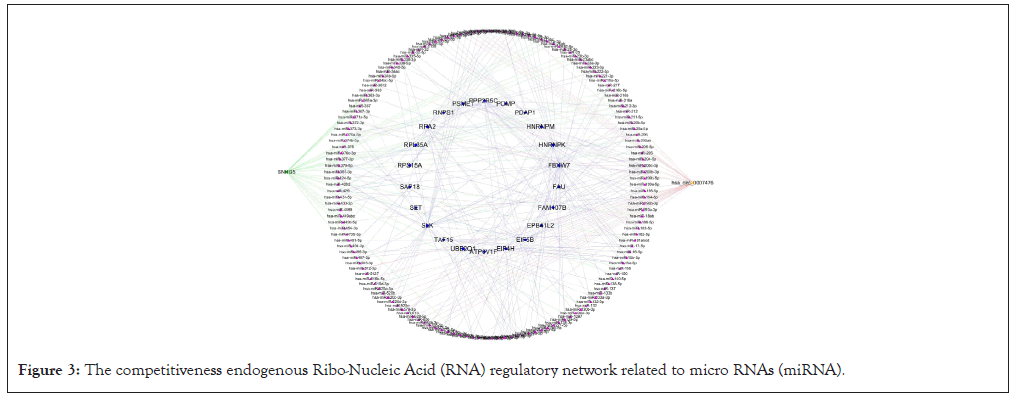
Figure 3: The competitiveness endogenous Ribo-Nucleic Acid (RNA) regulatory network related to micro RNAs (miRNA).
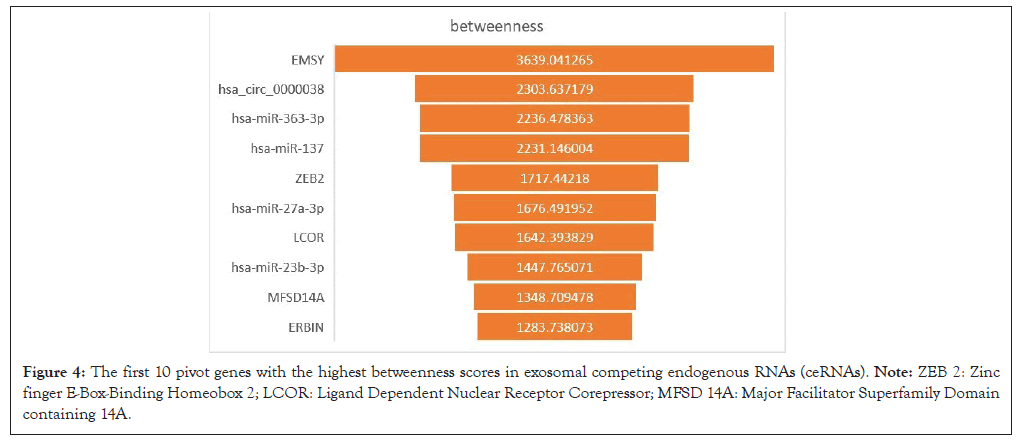
Figure 4: The first 10 pivot genes with the highest betweenness scores in exosomal competing endogenous RNAs (ceRNAs). Note: ZEB 2: Zinc finger E-Box-Binding Homeobox 2; LCOR: Ligand Dependent Nuclear Receptor Corepressor; MFSD 14A: Major Facilitator Superfamily Domain containing 14A.
GO annotation analysis
The Gene ontology of overlapping genes is as shown (Figure 5). The biological processes of differential exosomal mRNA included cytoplasmic translation, ribosome (nucleoprotein complex, large subunit), ribosomal Ribo-Nucleic Acid (rRNA) processing, rRNA metabolic process, ribosome (large subunit, ribonucleoprotein complex) assembly and non-coding RNA (ncRNA) processing. As we know, the occurrence of intestinal gastric cancer goes through a multi-step process, involving mutations such as Tumor Protein (P53) gene, while diffuse gastric cancer involves the reduction or deletion of cadherin, which is also detected in the biological processes involved in these differential genes, but not on the top 10 range [16-20]. The paths involved in the former are as exhibited (Table 1). The cell components where the genes located were mainly in the ribosome, its subunit (large and small subunit) and polyribosomes formed by ribosomes, focal adhesion, cell-substrate junction, proton-transporting Adenosine Triphosphatase (ATPase) (two-sector) and Adenosine Triphosphate (ATP) synthase complex (catalytic domain), endoplasmic reticulum membrane (its cytoplasmic side, rough endoplasmic reticulum and its membrane). Molecular functions included protein translation and modification, cadherin binding, rotational mechanism and proton channel activity, etc.
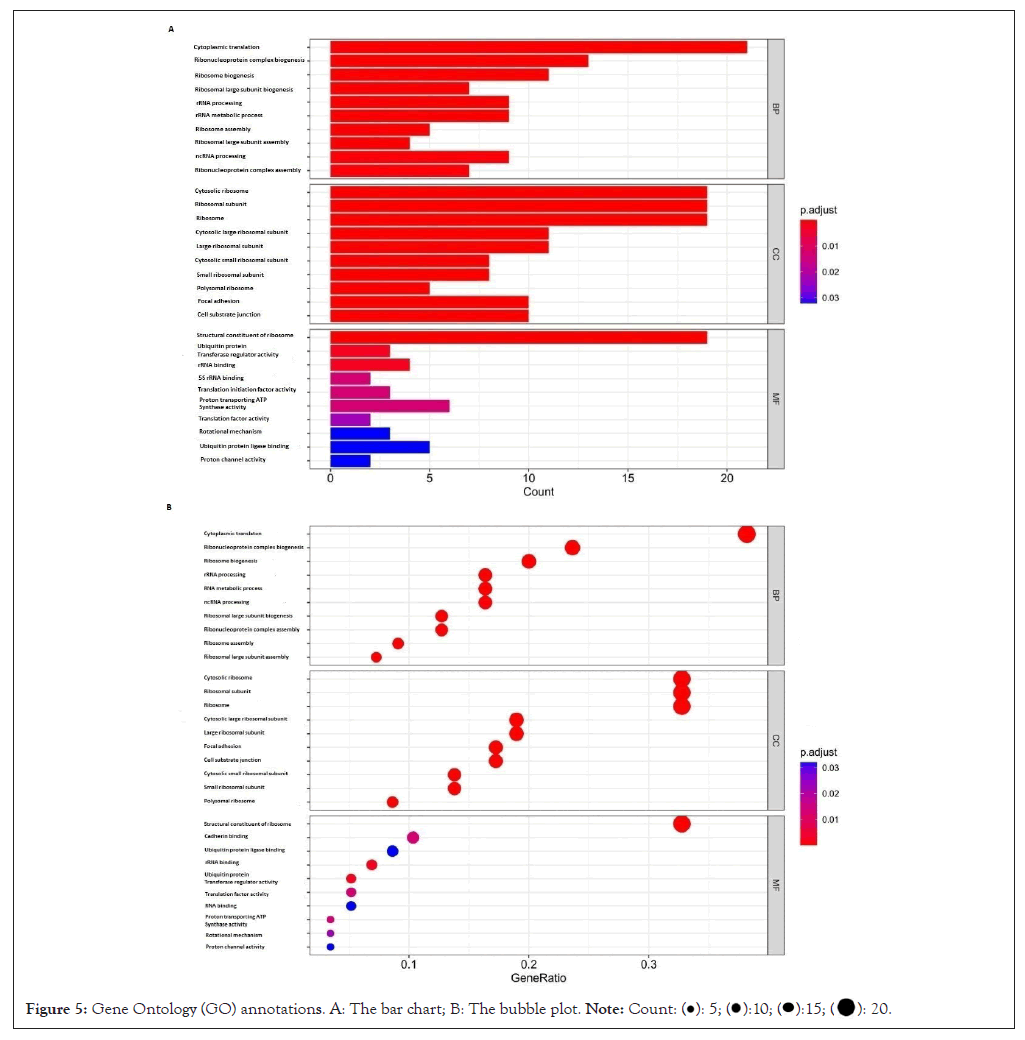
Figure 5: Gene Ontology (GO) annotations. A: The bar chart; B: The bubble plot. 
| Ontology | Description | p-value | Gene ID | Count |
|---|---|---|---|---|
| BP | Regulation of intrinsic apoptotic signaling pathway by p53 class mediator. | 0.000119539 | HNRNPK/RPL11/RPL26 | 3 |
| BP | Regulation of signal transduction by p53 class mediator. | 0.000246564 | HNRNPK/RPL11/RPL26/RPL5 | 4 |
| BP | Regulation of intrinsic apoptotic signaling pathway in response to Deoxyribo-Nucleic Acid (DNA) damage by p53 class mediator. | 0.001099219 | HNRNPK/RPL26 | 2 |
| BP | Signal transduction by p53 class mediator. | 0.001606558 | HNRNPK/RPL11/RPL26/RPL5 | 4 |
| BP | Intrinsic apoptotic signaling pathway by p53 class mediator. | 0.001693755 | HNRNPK/RPL11/RPL26 | 3 |
| BP | Positive regulation of signal transduction by p53 class mediator. | 0.003431736 | RPL11/RPL26 | 2 |
Note: BP: Biological Process; p53: Tumor Protein.
Table 1: The paths involved in the p53 gene.
Enrichment analysis of KEGG
The KEGG pathway shows that the DEmRNA is mainly concentrated in the mRNA monitoring pathway (involving mRNA Protein Phosphatase-2 Regulatory Subunit B'Gamma (PPP-2 R5C), RNA-binding Protein with Serine-rich Domain 1 (RNPS1), Sin3A Associated Protein 18 (SAP18)), proteasome (involving mRNA, Proteasome Maturation Protein (POMP) and Proteasome Activator Complex Subunit 1 (PSME 1)), ribosome (assuming mRNA FAU, RPL35A and RPS15A), as shown (Figure 6).
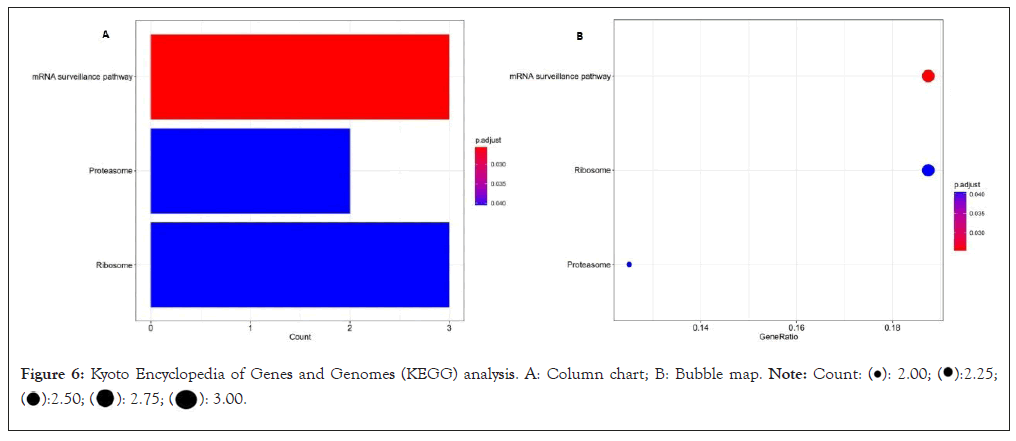
Figure 6: Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. A: Column chart; B: Bubble map. 
Gastric Cancer (GC) is one of most often malignancy of the alimentary system. The mortality of GC is high and the prognosis is unsatisfactory because of the detection often in the late stage of diagnosis [21]. Therefore, it is crucial to reduce the incidence rate and mortality of GC by early detection and the use of effective screening methods [22,23].
Exosomes are small vesicles excreted by living cells, with typical lipid bilayer membrane, carrying a variety of meaningful message such as proteins, lipids, nucleic acids, etc., play a significant role in the transmission of intercellular substances and information and gradually become a biomarker for early diagnosis of many diseases [24-26]. Nucleic acids include DNA and RNA (mRNA, miRNA, lncRNA and tRNA), which provides a good data source for us to establish a competitive endogenous regulatory network of multiple RNAs [27,28].
In this study, 62 DEmRNA, 3 DElncRNAs and 1 DEcircRNAs were searched out and the ceRNA system was built with their binding miRNA prediction data. Then, first 10 pivot genes with the highest betweenness scores were distinguished by cytoscape software including 5 mRNAs (EMSY, ZEB 2, LCOR, MFSD 14A and ERBIN), 4 miRNAs (hsa-miR-363-3p, -137, -27a-3p and -23b-3p) and 1 circRNA (hsa_circ_0000038). The GO annotation and the enrichment analysis of KEGG pathway showed that ribosome biogenesis played a major role in the exosome of gastric cancer.
It is reported that ZEB 2 expression was be relevant to the efficacy of platinum chemotherapy and the miR-338-5p-ZEB2 axis has diagnostic and prognostic value [29]. ZEB 2, as an epithelial mesenchymal transformation regulator, inhibited by miR-200c, could inhibit the metastasis of non-small cell lung cancer [30]. The highly expressed lncRNA ZEB 2 Antisense RNA 1 (ZEB 2 AS1) in GC patients had a poor prognosis [31]. The high expression of LCoR was considered as a poor prognosis marker in GC [32]. ERBB2 Interacting Protein (ERBB2IP) was upregulated gene, which might be in connection with the resistance to intraperitoneal paclitaxel in GC with peritoneal metastasis [33]. Hsa-miR-363-3p was probably a therapeutic targets and down-regulated in Tibetan with GC [34]. MiR-137 was negative correlation with Enhancer of Zeste Homolog 2 (EZH 2) and its expression was down-regulated in GC [35]. Hsa-miR-27a-3p might serve critical roles in early GC [36]. However, the role of EMSY, MFSD 14A, hsa-miR-23b-3p and hsa_circ_0000038 in the development and diagnosis of GC were still unclear, which could be used as potential targets for future research.
To sum up, this study used bioinformatics methods to identify exosomal DeRNA (mRNA, lncRNA and circRNA) related to GC occurrence, constructed corresponding ceRNA networks and speculate that ribosome biogenesis played a major role in the exosome of gastric cancer, which provided a molecular basis for further research on GC progress and metastasis and provided new liquid biopsy biomarkers for early diagnosis of GC. However, due to the small sample size of disease exosomes, further clinical multicenter validation is required. Different types of gastric cancer, different pathogenesis of adherent cancer and non-adhesive cancer and their differences in ceRNA also need to be clarified.
Ming-ming He and Yuan Zhong: Data collection, data analysis, manuscript writing. Tao Lei, You-li Jian and Xian-kui Cheng: Data analysis. Chun-yan Lv: Project development and critical revision of manuscript.
This study was sustained by the Department of Science and Technology Key R and D projects in Sichuan Provincial (grant number: 2022YFS0200).
At the point of finishing this paper, I'd like to express my sincere thanks to all those who have lent me hands in the course of my writing this paper. I'd like to thank those leaders, teachers and working staff especially those in Chengdu University. Without their help, it would be much harder for me to finish my study and this paper.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Lv C, Cheng X, Zhong Y, Jian Y, He M, Lei T (2024) Ribosome Biogenesis: A Major Player in the Exosome of Gastric Cancer. J Proteomics Bioinform. 17:663.
Received: 14-Feb-2024, Manuscript No. JPB-24-29593; Editor assigned: 16-Feb-2024, Pre QC No. JPB-24-29593 (PQ); Reviewed: 01-Mar-2024, QC No. JPB-24-29593; Revised: 08-Mar-2024, Manuscript No. JPB-24-29593 (R); Published: 15-Mar-2024 , DOI: 10.35248/0974-276X.24.17.663
Copyright: © 2024 Lv C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.