Autism-Open Access
Open Access
ISSN: 2165-7890
ISSN: 2165-7890
Research Article - (2024)Volume 14, Issue 2
Introduction: Autism Spectrum Disorder (ASD) is a group of developmental disorders with multifactorial genetic and environmental etiology. FOXP1 gene product plays an important role in the development of Central Nervous System (CNS) tissues and neuron function. The role of mutations of certain genes as cause of ASD was highly investigated. One of the blamed gene is Single Nucleotide Polymorphism (SNP) of FOXP1 rs112795301 in association with ASD that represents a fruitful area of research.
Aim: To unveil the frequency distribution of FOXP1 SNP rs112795301 and whether it has a role in pathogenesis of ASD patients.
Materials and Methods: A case-control study was done in Faculty of Medicine, University of Kufa from December, 2019 to December, 2020. We determined SNP rs112795301 of FOXP1 genes using allele specific Polymerase Chain Reaction (PCR) technique 60 autistics vs. 60 control group. Odd ratios, 95% Confidence Interval (CI) and the Chi-square test were employed to assess the genotype and allele distributions in patient groups. Statistical Package for Social Sciences (SPSS) were utilized for all statistical analysis.
Results: The analysis of frequency distribution of allele genotypes of SNP rs112795301 in FOXP1 gene (autistic group vs. control group) showed that high significant association of AA genotype, Odds ratio (OR) 4.2759 (genotype AA four times more in patient than in control) and GG genotype more with control. The A allele more in patient OR 2.6337 but no significant difference in relation to both gender and disease severity in genotyping and allelic distribution.
Conclusion: Variants of FOXP1 gene SNP rs112795301 are associated with ASD in Iraqi population, which is likely role of SNPs in etiology ASDs.
Autism spectrum disorder; FOXP1 gene; SNP rs112795301
Autism Spectrum Disorders (ASDs) are complex multifactorial neurodevelopmental disorders. It is a condition defined by three basic deficiencies in social interaction and communication, as well as constrained repetitive and stereotyped patterns of behavior, interests and activities, which is not a disease, but rather a syndrome with various epigenetic and genetic origins [1]. The FOXP1 gene belongs to the Forkhead box (FOX) transcription factor family's subfamily P that acts as a repressor of transcription, it plays a significant role in the regulation of type-specific gene transcription [2-4]. In mammals, FOXP1 controls a number of essential facets of development, including lung, brain, thymus and heart tissue development also involved in esophageal muscle growth and epithelial development [5,6]. FOXP1 is abundant in the neocortex, hippocampus and striatum during development and maturation. Several studies have begun to explain FOXP1 involvement in the brain. FOXP1 mutations have been seen in a small number of human patients like intellectual disabilities and autism spectrum disorder, as well as language deficiency and Developmental Verbal Dyspraxia (DVD) [7].
FOXP1 heterozygous deletions, whole gene deletion, gene with repeated de novo mutations, translocations, nonsense variations, mis-sense variants and frameshift variants have all been found in individuals with ASD and/or Intellectual Disability (ID) [8-13]. The discovery of whole gene deletions shows that haplo insufficiency is the pathogenic mechanism [14]. The appearance of speech and language deficits as a symptom of FOXP1 deficient syndrome and damaged in case of linguistic dysfunction conditions.
Study population
During the period from December 2019 to December 2020, a total of 60 patients with a median age of 9 years (range 3-15) years 6.66 ± 3. 051(mean ± Standard Deviation (SD)), 45 males and 15 females, who were clinically diagnosed with ASD by a consultant psychiatrist, healthy control group who were matched in age and gender were subjected to the present study.
The present study was done in Faculty of Medicine, University of Kufa after getting the required bioethical approval from the Institutional Review Board (IRB) committee of the college. All the cases which were included in the present study attended the teaching hospitals which are located in Middle and South Euphrates Regions, Iraq. Families who expressed interests in the study were signed a formal written consent sheets and given a questionnaire to complete.
Selection of the ASD cases
Patients who fulfilled the criteria of the Diagnostic and Statistical Manual of Mental Disorders (DSMMD) (5th edition) being selected as ASD subject. The diagnosis was carried out by a psychiatrist. Patients with typical ASD were selected to be enrolled in the study after taking the consent from their parents. The patients were split into three groups based on whether they met mild autistic people (n=39), moderate autistic people (n=13) and severe autistic people (n=8). Young controls were generally developing, healthy, unrelated to the individuals who had autism and they did not match any of the exclusion criteria included the following:
• Rett syndrome, focal epilepsy or other disorders that might be a cause of ASD.
• Associated condition involving lesion above the brain stem.
• History of brain damage.
• Congenital neurologic diseases.
• Present or past history of severe visual or auditory impairment.
• Severe malnutrition or psychological deprivation.
Blood sampling and processing
Blood sample was obtained from both healthy control and ASD patients in parallel. About 2.5 ml of venous blood were collected. Blood samples were stored in Ethylene Diamine Tetra Acetic Acid (EDTA) tube to be used later for Deoxyribonucleic Acid (DNA) extraction and gene analysis by PCR techniques.
Isolation of genomic DNA
Following the manufacturer's instructions, genomic DNA was extracted from blood samples using the Bosphore® genomic DNA extraction spin kit. The nano-drop spectrophotometer was used to evaluate the genomic DNA that had been extracted. DNA concentrations were determined at a 260 nm wavelength and expressed in ng/L. Agarose gel electrophoresis was carried out according to the methods detailed by Sambrook and Fritsch. Single Nucleotide Polymorphisms (SNP) were genotyped in the study cohorts. The SNPs of accession numbers SNP rs112795301 in FOXP1 gene were genotyped in all study participants using allele specific PCR technique.
Allele specific PCR primer
PCR primers utilized in this study were allele specific. The suspect SNPs were briefly retrieved from the dbSNP database, which can be found on the server https://www.ncbi.nlm.nih.gov/snp/, it contains human single nucleotide polymorphism variations, microsatellites, small scale-insertions and deletions. Most frequently, a nucleotide sequence of 1000 base pairs (bp) or 500 bp containing the dubious SNP. Then, using the primer-blast online program, which is located at the server (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) from National Center for Biotechnology Information (NCBI), the allele specific primers were manually designed and computationally checked regarding 3′ complementarity, 3′ self-complementarity, GC content and melting temperature. In this work, all designed primers were created using Integrated DNA Biotechnology (IDT Co., Canada). Genotyping of the SNPs with accession numbers rs112795301 in the FOXP1 gene was done using DNA from study participants and an allele-specific PCR method. Each DNA sample underwent two PCR procedures tailored to its particular allele. A primer pair consisting of a forward primer bearing the allele SNP base at the 3′ prime end and a reverse primer was used to direct each allele-specific PCR reaction. The rules were followed when reading the results. The allele-specific primer sequences and PCR conditions for the FOXP 1 SNPs are given.
Statistical analysis
Numerical data from the current study were represented as mean ± SD. Results were statistically analyzed by using Statistical Package of Social Sciences (SPSS) version 27. Statistical difference between two means and multiple groups’ means were tested using student t-test and one-way Analysis of Variance (ANOVA), respectively. Regarding genomic analysis, the representations of genotypes and alleles were determined by the Hardy-Weinberg Equilibrium (HWE). This is done by making a comparative analysis between what is expected and the observed allele frequencies. In order to estimate genotype and allele frequencies among patients and controls, chi-square test was used. To analyze the frequency distribution of genotypes and alleles, Odds Ratios (OR) with 95% confidence intervals (95% CI) were determined. Basically, a p-value of <0.05 was considered statistically significant at 95% CI, while a p-value of <0.001 was considered highly significant.
Characteristics of the patient
The median age of ASD patients was 6.66 with a range of 3 to 16, while the median age of controls was 5.76 with a range of 3 to 16. The ratio of males to females was substantially larger (p>0.05) in the ASD (n=60) and matching healthy control populations (n=60). In both, ASD and control, the consanguinity percentage was high, at 36.66% and 40%, respectively (Figures 1a-1d).
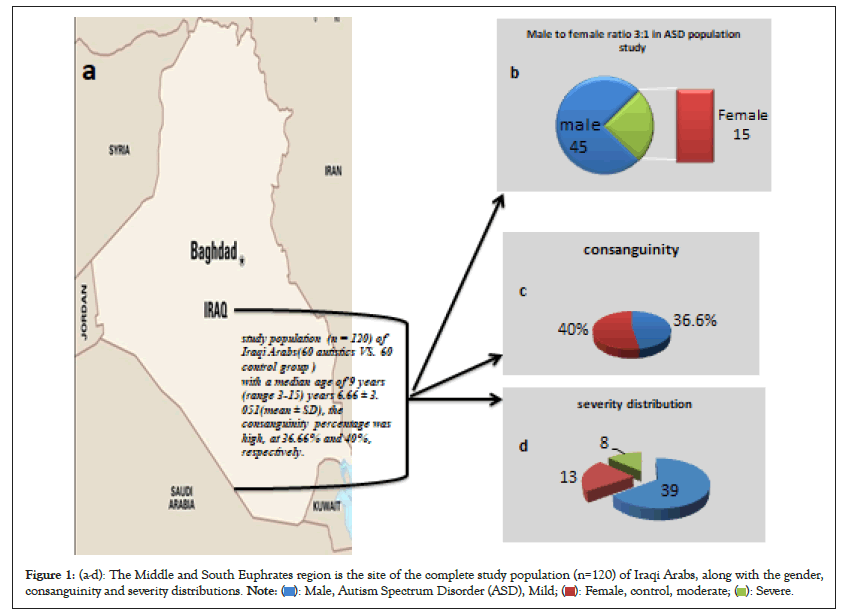
Figure 1: (a-d): The Middle and South Euphrates region is the site of the complete study population (n=120) of Iraqi Arabs, along with the gender,
consanguinity and severity distributions. 
According to Diagnostic and Statistical Manual of Mental Disorders (DSM-V) severity categorization, the details of the demographic of the patients with ASD are shown in Table 1. The majority of ASD patients (n=39) were classified as having mild autism symptoms. Children of consanguineous marriages (35.89%; 95% CI 0.41180-2.2381, OR 0.967) and recurrent in the same family 15.38%. Four of the 13 patients with intermediate severity (30.76%; 95% CI 0.2114-2.7876, OR 0.767) were the offspring of consanguineous unions and recurrent in the same family was 07.69%. In patients with severe ASD categorization (n=8, 50%; 95% CI 0.3924-7.6035, OR 1.727), the odds ratio for consanguinity was highest but, no recurrent in same family (Table 1).
| Groups | Number | Consanguinity | Unrelated | OR | P value | 95% CI | Recurrent in same family |
|---|---|---|---|---|---|---|---|
| ASD | 60 | 22 (36.66) | 38 (63.33) | 1 | 1 | 0.4759-2.1014 | 7 (11.66) |
| Mild ASD | 39 | 14 (35.89) | 25 (64.1) | 0.967 | 0.938 | 0.4180-2.2381 | 6 (15.38) |
| Moderate ASD | 13 | 4 (30.76) | 9 (69.23) | 0.767 | 0.687 | 0.2114-2.7876 | 1 (07.69) |
| Severe ASD | 8 | 4 (50) | 4 (50) | 1.7273 | 0.469 | 0.3924-7.6035 | 0 (0) |
Note: ASD: Autism Spectrum Disorder; OR: Odds Ratio; CI: Confidence Interval.
Table 1: Demographic characteristics of the autistic and controls.
ASD and control groups' SNP rs112795301, genotype and allele distributions for the FOXP1 gene. Data in Table 2 reveal the pattern of SNP rs112795301 in foxp1 gene among the target samples in this study reflecting the three possible genotypes raised in the sample (GG/GA/AA). Figure 2 shows gel electrophoresis detection of the results A and G allele (Table 2).
| Genotype/allele | Patients (N=60) | Control (N=60) | P value | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | ||||
| Genotype | |||||||
| GG | 18 | 30 | 28 | 46.66 | 0.062 (NS) | 0.4898 | 0.2314 to 1.0366 |
| GA | 11 | 18.33 | 20 | 33.34 | 0.007 (S) | 0.322 | 0.1403 to 0.7390 |
| AA | 31 | 51.67 | 12 | 20 | 0.0004 (S) | 4.2759 | 1.9016 to 9.6145 |
| Alleles | |||||||
| G | 47 | 39.17 | 78 | 65 | 0.0002 (S) | 0.3797 | 0.2265 to 0.6366 |
| A | 73 | 60.83 | 46 | 35 | 0.0002 (S) | 2.6337 | 1.5707 to 4.4159 |
Note: (NS): No significant association (P>0.05); (S): Significant association (P<0.05); OR: Odds Ratio; CI: Confidence Interval; No.: Number of patients.
Table 2: Distribution of the genotypes (GG, GA, AA) and alleles (G, A) of FOXP1 gene in study population.
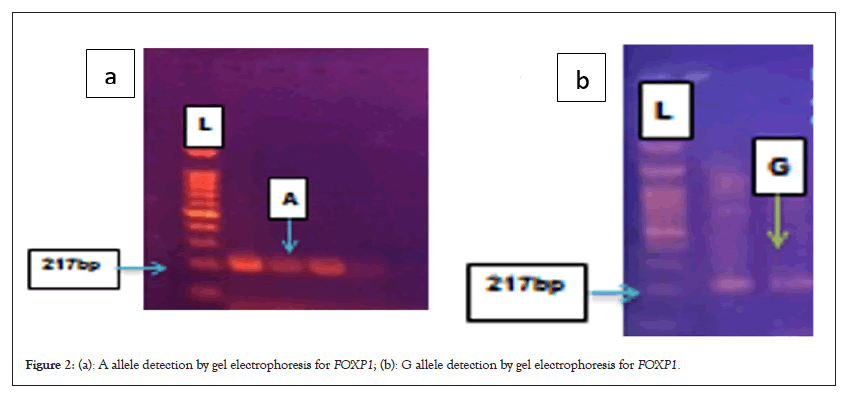
Figure 2: (a): A allele detection by gel electrophoresis for FOXP1; (b): G allele detection by gel electrophoresis for FOXP1.
The genotype AA is four times more prevalent in patients in comparable to controls, whereas the genotype GG is more prevalent in control samples, which indicating a strong significant connection with AA genotype. The patients A, allele is more prevalent OR 2.6337. For allele distribution, high significant for both alleles however, the A allele is more in patients (Figures 2a and 2b).
Genotype and allele distribution of SNP rs112795301 according to gender
The genotype and allele distribution of SNP rs112795301 in FOXP1 gene according to the gender was studied among the autistic group Table 3. Data reveals no significant difference in GG/GA/AA genotypes for SNP rs112795301 in FOXP1 gene nor for the alleles G/A among the autistic patients (Table 3).
| Genotype/allele | Male (N=45) | Female (N=15) | DF | χ² | P-value | ||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | ||||
| Genotype | |||||||
| GG | 12 | 26.67 | 6 | 40 | 1 | 0.952 | 0.329 (NS) |
| GA | 10 | 22.22 | 1 | 6.66 | 1 | 1.81 | 0.178 (NS) |
| AA | 23 | 51.11 | 8 | 53.34 | 1 | 0.022 | 0.881 (NS) |
| Alleles | |||||||
| G | 34 | 37.78 | 13 | 43.34 | 1 | 0.291 | 0.589 (NS) |
| A | 56 | 62.22 | 17 | 56.66 | 1 | 0.291 | 0.589 (NS) |
Note: (NS): No significant association (P>0.05); DF: Deviation Factor; χ²: Distribution number; No.: Number of male and female patients.
Table 3: Distribution of SNP rs112795301 in foxp1 genotypes and alleles among ASD group in relation to gender.
Distribution of SNP rs112795301 in foxp1 genotypes and alleles among ASD group in relation to severity
The distribution of the genotypes and alleles for SNP rs112795301 in foxp1 gene according to the severity was studied among the ASD groups with its three stratified patients according to the severity of the symptoms. Data depicted in Table 4 reveals no significant difference neither in the genotypes nor in the alleles among the three stratified ASD patients with outcomes mild, moderate and severe (Table 4).
| Genotype/allele | Mild (N=39) | Moderate (N=13) | Severe (N=8) | DF | χ² | P-value | |||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||||
| Genotype | |||||||||
| GG | 10 | 25.64 | 4 | 30.77 | 4 | 50 | 2 | 1.88 | 0.391 (NS) |
| GA | 60 | 15.38 | 4 | 30.77 | 1 | 12.5 | 2 | 1.75 | 0.417 (NS) |
| AA | 23 | 58.98 | 5 | 38.46 | 3 | 37.5 | 2 | 2.38 | 0.304 (NS) |
| Alleles | |||||||||
| G | 26 | 33.33 | 12 | 46.16 | 9 | 56.25 | 2 | 3.6 | 0.165 (NS) |
| A | 52 | 66.67 | 14 | 53.84 | 7 | 43.75 | 2 | 3.6 | 0.165 (NS) |
Note: (NS): No significant association (P>0.05); DF: Deviation Factor; χ²: Distribution number; No.: Number of Autism Spectrum Disorder (ASD) patients with mild, moderate and severe condition.
Table 4: Genotype and allele distribution of SNP rs112795301 in foxp1 gene among ASD group in accordance with disease severity.
Data from the current study reveal that AA genotype of Forkhead-box Protein P1 (FOXP1), rs112795301 is more frequent associated with patient with ASD; this could be a pathogenic in the development of ASD. On the other hand, GG genotype is more common with control group which seems to be protective [15].
The finding that genetic mutation of FOXP1 is related to ASD is consistent with many previous studies. It has been reported that the gene haplo insufficiency of the Forkhead-box Protein P1 (FOXP1) play a vital role in the causation of many neurodevelopmental disorder including, intellectual disability, developmental delay, speech deficits, ASD and many other neurodevelopmental disorder traits Lozano, et al., [5], Sollis, et al., [16]. In fact, a large amount of data has identified many FOXP1 gene mutations including, point mutations, duplications and heterozygous deletions and though as casual for autism. Furthermore, number of mutations in FOXP1 gene has shown to be associated with ASD [17]. Bacon, et al., [4], who reported altered social behavior, striatal morphology and hippocampal electrophysiology in mice which underwent a brain specific FOXP1 loss. It has been reported, the deletions in FOXP1 resulting loss of function of FOXP1 gene that was sufficient enough to produce many neurodevelopmental disorder including ASD, speech impairment and other dysmorphic features [18].
Tang B and his co-workers reported that FOXP1 was functioning as a transcriptional repressor of the immune signaling in mouse brain, FOXP1 played a role in the pathophysiology of Huntington disease. They suggested the any elevation in FOXP1 gene expression in Lymphoblastoid Cell Line (LCL) of autistic patients may provide much newer insight which dysregulated immune signaling in the brain which motivate ASD. In another relevant case series study detected three subjects who were suffering from mental retardation, speech deficits, two with autistic features who have heterozygous deletion overlapping FOXP1 gene [19].
The current study revealed no significant difference in the genotypes GG/GA/AA for SNP in relation to the gender. However, until recently, there is a paucity of finding in the literature considering this relation. Considering correlation of SNP rs112795301 in foxp1 gene with disease severity, the current study revealed that no significant difference neither in the genotypes nor in the alleles with ASD disease severity. Again, there is limited information in the literature about this finding. It is recommended to further investigate to correlation of FOXP1 SNP with gender of patients with ASD and disease severity.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Al-Ali ZA, Al-Mousawi MR, Al-Karaqully AJ, Alattabi AS, Ali MS, Mahdi LH, et al (2024) Role of FOXP1 (SNPS) Gene in Autism Spectrum Disorder Pathogenesis. Autism-Open Access. 14:397
Received: 31-May-2024, Manuscript No. AUO-24-30077; Editor assigned: 03-Jun-2024, Pre QC No. AUO-24-30077 (PQ); Reviewed: 17-Jun-2024, QC No. AUO-24-30077; Revised: 24-Jun-2024, Manuscript No. AUO-24-30077 (R); Published: 02-Jul-2024 , DOI: 10.35248/2165-7890.24.14.397
Copyright: © 2024 Al-Ali ZA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.