Journal of Leukemia
Open Access
ISSN: 2329-6917
ISSN: 2329-6917
Research Article - (2019)Volume 7, Issue 1
Abstract: Th17 cells (subset of CD4+ cells) are characterized by interleukin (IL)-17A and IL-17F production, which share strong homology, and surface expression of the IL-23 receptor (IL-23R). They have been implicated in the pathogenesis of inflammatory, autoimmune diseases and in many different types of human tumors, including lymphoma and myeloma.
Objectives: The aim of this study was to determine the association between the polymorphic features located within the IL-17A, IL-17F and IL-23R genes and susceptibility to or association with AML and the relationship between these polymorphic variants and the plasma IL-17 levels in all studied groups.
Subjects and Methods: 82 individual of Egyptian population including 27 AML patients, 40 individual who are relatives of those patients and 15 apparently healthy controls. All were subjected to detection of Interleukin 17F gene polymorphism by 2 steps: first Conventional Polymerase Chain Reaction then Restriction Fragment length Polymorphism by Restriction enzymeHIN1II (NLAIII) (PCR-RFLP) assay, Detection of Interleukin 17A and Interleukin23 receptor genetic polymorphisms by real time Polymerase Chain Reaction and Quantification of Interleukin 17 gene protein product by Enzyme Linked Immunosorbent Assay (ELISA).
Results: our results revealed that the (G) variant of IL17F and its homozygosity were significantly higher among patients compared to relatives and healthy individual. While IL17-A and IL23-R polymorphisms elicit non statistical significant difference among the three groups. Also a significant correlation was observed between IL17 protein plasma level and the three polymorphisms (IL17F, IL17A and IL23R genetic polymorphisms).
Conclusions: IL-17F gene G single mutant and GG homozygous mutant were associated with acute myeloid leukemia susceptibility in Egyptian population while IL17-A and IL23-R polymorphisms were not associated with susceptibility to the disease. Also there were statistical significant association (PË0.05) and high statistical significant (PË0.001) association between ELISA (IL17 plasma level) results and the three genetic polymorphisms in studied groups. There was high statistical significant difference (PË0.001) between patients and other two groups in IL17 plasma level by ELISA.
Acute myeloid leukemia; Leukocytosis; Interleukin
Acute Myeloid Leukemia (AML) is a life-threatening hematopoietic stem cell neoplasm characterized by bone marrow infiltration by leukemic cells that suppress normal hematopoiesis, frequently resulting in fatal infection, bleeding or organ infiltration, with or without leukocytosis [1,2]. It is the most common acute leukemia that affects adults, and its incidence increases with age. Of those diagnosed at a later age, the diagnosis is often associated with underlying Myelodysplastic Syndromes (MDS), sometimes linked to cancer chemotherapy and radiotherapy exposure [3,4]. The etiology of AML is heterogeneous and complex, both environmental and genetic factors play significant roles in the development of AML [5]. AML has several subtypes with different treatment protocol and prognosis. AML cure and average survival didn’t reach satisfactory rate, so extended work on AML patients is needed [6].
It has been reported that the Th17 cell frequencies or levels of its product Interleukin-17 (IL-17) and its related cytokines might be involved in AML pathogenesis [7,8]. The hallmark of the Th17 subset is the production of interleukin17F (IL-17F) and Interleukin17A (IL-17A), which share strong homology, and surface expression of the Interleukin23 receptor (IL-23R). IL-23R is essential for the differentiation of Th17 cells and plays a key role in the development of the pathogenic Th17 cells which produce the cytokine IL-17 [9].
Th17 cells producing IL17 are important in host defense against microbial infections, including bacteria, mycobacteria, viruses, and parasites [10,11]. They also appear to be key mediators in the pathogenesis of a broad array of inflammatory and autoimmune diseases, including rheumatoid arthritis, psoriasis, and inflammatory bowel disease [10].
IL17A (IL17) is a pro-inflammatory cytokine that acts on a variety of cells (e.g., fibroblasts, epithelial cells, endothelial cells, and monocytes) to induce the production of other cytokines, including IL6, tumor necrosis factor-α (TNFα), Granulocyte-Macrophage Colony-Stimulating-Factor (GMCSF), Granulocyte Colony- Stimulating-Factor (GCSF), chemokines (chemokine (C-X-C motif) ligand 1 (CXCL1), CXCL2, CXCL5, and CXCL8), antimicrobial peptides (defensins) and matrix metalloproteinases (MMP1, MMP3 and MMP13). These factors mediate the recruitment, activation and migration of neutrophils and myeloid cells, and also induce angiogenesis and tissue destruction [12,13].
Th17 cells producing IL17 have been implicated in many different types of human tumors, including lymphoma [14], myeloma [15,16], breast cancer [17,18], colon cancer [18-20], gastric cancer [21,22] hepatocellular cancer [19,23], melanoma [18,19,24], ovarian cancer [19,25-28] pancreatic cancer [19], and prostate cancer [29,30].
The role of IL17 in susceptibility to tumors occurs by two suggested mechanisms. First mechanism is chronic infection and inflammation which are clearly important factors for tumorgenesis. Second suggested mechanism is that tumor cells, as well as tumor-derived fibroblasts, secrete MCP-1 (the ligand for CCR2 or CCR4; alias CCL2) and RANTES (the ligand for CCR1, CCR3 or CCR5; alias CCL5), both of which strongly attract Th17 cell migration [18].
The aim of this study was to determine the association between the polymorphic features located within the IL-17A, IL-17F and IL- 23R genes and susceptibility to or association with Acute Myeloid Leukemia (AML) in Egyptian population and the relationship between these polymorphic variants and the plasma IL-17 levels in all studied groups.
The present study was conducted in clinical pathology department, AL-Azhar and Zagazig University hospitals. The study was performed on 82 individuals collected in the period from October 2016 to May 2018. They were classified into 3 groups: Group I: Included 27 patients (14 males and 13 females) suffering from Acute Myeloid Leukemia (AML). Their ages ranged from 16 y to 72 y. They are divided into subgroups according to type of AML: Sub group 1: M1 & M2 (12 cases), Sub group 2: M3 (4 cases), Sub group 3: M4 (3 cases), Sub group 4: M5 (6 cases) and Sub group 5: Bilineage leukemia (2 cases), Group II: Included 40 individual. They are relatives of the AML patients (22 males and 18 females). Their ages ranged from 15 y to 70 y and Group III: included 15 apparently healthy individual (9 males and 6 females). Their ages ranged from 25 y to 70 y. All studied groups were diagnosed at clinical pathology department at AL-Azhar and Zagazig University hospitals. They were matched as regard age and sex. Patients received chemotherapy or drugs or radiotherapy before and patients have previous solid tumors or non-myeloid leukemia was excluded. All studied groups given their written, informed consent to participate.
All patients, relatives and controls were subjected to:
1. Full history taking and clinical examination
2. Laboratory investigation which include:
• Complete Blood Count (CBC) including Leishman-stained Peripheral Blood (PB) smears examination,
• Bone marrow aspiration with examination of Leishman-stained smears (for patients),
• Detection of Interleukin 17F gene polymorphism by 2 steps: first step is Conventional Polymerase Chain Reaction; second step is R estriction Fragment length Polymorphism by Restriction enzyme HIN1II (NLAIII) (PCR- RFLP) assay,
• Detection of Interleukin 17A and Interleukin23 receptor genetic polymorphisms by real time Polymerase Chain Reaction,
• Quantification of Interleukin 17 gene protein plasma level by Enzyme Linked Immunosorbent Assay (ELISA).
Four mL venous blood samples were withdrawn from each individual participating in this study under complete aseptic conditions. Samples were divided into: 1.0 mL on plain vaccutainer for ELISA determination, 1.0 mL on sterile EDTA vaccutainer for Complete Blood Count (CBC) and 2.0 mL on sterile EDTA vaccutainer for genetic studies of IL-17F, IL-17A and Interleukin23 receptor genetic polymorphisms.
• Blood Genomic DNA extraction Protocol for all samples undergoing genetic studies [31].
• Detection of 17F gene polymorphisms by restriction fragment length polymorphism (RFLP) [32,33].
a) Amplification by conventional PCR [32]. Primer 1 (forward) (5 end to 3 end) (GTT CCC ATC CAG CAA GAG AC), Primer 2 (reversed) (3 end to 5 end) (AGC TGG GAA TGC AAA CAA AC) and the incubation for 2 hours in thermal cycler for (40 cycles). The PCR conditions were as follow: initial denaturation at 94°C for 3 min, denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, extension at 72°C for 30 sec and final elongation at 72°C for 7 min then read on gel electrophoresis (120 V for 30 min).
b) Restriction fragment length polymorphism by restriction enzyme HIN1II (NLAIII) 33. Then we incubated samples for 2 hours at 37°C then read on gel electrophoresis (120 V for 30 min).
• Detection of 17F protein in patient plasma by ELISA (Enzyme- Linked Immunosorbent Assay) by IL-17 Cytoscreen kit [34].
• Detection of Interleukin 17A and Interleukin 23 receptor genetic polymorphisms by Real time PCR [35-37]. The plate inserted into Step one analyzer with Standard protocol: Pre PCR read step 1 at 600C for 30 sec, Initial enzyme activation step 2 at 95°C for 10 min, Denaturation step 3 at 95°C for 15 sec, Annealing and extension step 4 at 60°C for 1min and Post PCR step 5 at 60°C for 30 sec then after 60 cycle read result. Two primers for amplifying the polymorphic sequence of Interest (Two TaqMan MGB probes for distinguishing between the two alleles, each TaqMan MGB probe contains A reporter dye at the 5′ end of each probe: VIC dye is linked to the 5′ end of the Allele 1 probe and FAM dye is linked to the 5′ end of the Allele 2 probe).
SPSS software version 25.0 (IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp, 2017) was used to analyze the data. Data are presented in tables and figures. Continuous data was presented as median and inter-quartile range (IQ range).
We used Kolmogorov-Smirnov and Levene tests to determine the distribution characteristics of variables and variance homogeneity. Qualitative data was presented as frequencies and proportions. Chi square test (χ2) and fisher’s exact test were used to analyze qualitative independent data. Kruskal-Wallis test (KW) and post hoc test was used to test differences when more than two independent groups were present.
The sensitivity, specificity and accuracy of ELISA in diagnosis of leukemia. Receiver Operating Characteristic (ROC) curves were plotted for the optimal cut-off values of ELISA that was predictive of leukemia as well as the sensitivity and specificity. The optimal cut-off values were defined as the values that allow discrimination between leukemia patients from relatives and control with highest sensitivity and specificity. In all the tests, p-value of <0.05 was taken as significant and <0.001 was taken as highly significant.
Table 1: ELISA (IL17 plasma level) results in studied groups.
| IL17 plasma level by ELISA | 1Patients group | 2Relatives group | 3Control group | KW | P | Post-hoc |
|---|---|---|---|---|---|---|
| IL17 (pg/ml): Median IQ-Range |
9.3 9.3–16.0 |
5.5 2.0–9.2 |
3.0 0.03–9.3 |
30.4 | <0.001 HS |
0.0001 0.0002 0.3943 |
1Patients group versus Relatives group, 2Patients group versus Control group and 3Relatives group versus Control group. This table shows that there was high statistical significant difference (p<0.001) between patients group and other two groups in IL17 plasma level results by ELISA. However, there was no statistical significant difference between relatives group and control group in IL17 plasma level results by ELISA (Table 1).
This table shows that, best cutoff point was ≥ 9.3 pg/mL with sensitivity and specificity (85.2% & 74.5%), Positive and Negative Predictive values (62.2% & 91.1%), accuracy (78.0%). Area under Curve was 0.87 (0.79-0.94) (Table 2 and Figure 1).
Table 2: Diagnostic performance of ELISA in diagnosis of AML.
| Cutoff point | AUC (95%CI) | Sensitivity | Specificity | PPV | NPV | Accuracy | P |
|---|---|---|---|---|---|---|---|
| ≥ 9.3 pg/ml | 0.87 (0.79–0.94) |
85.2% | 74.5% | 62.2% | 91.1% | 78.0% | <0.001 HS |
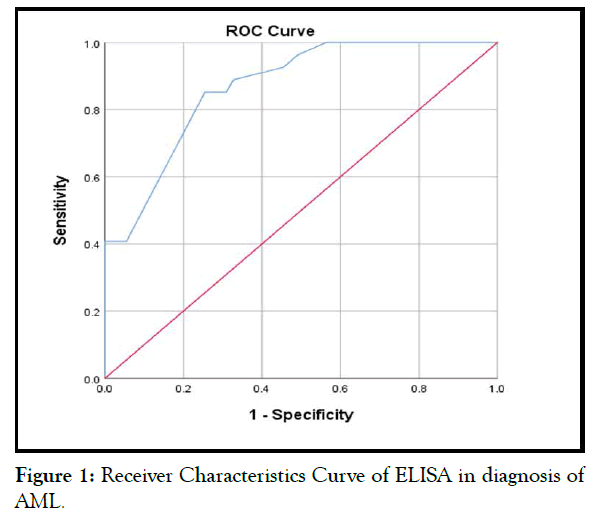
Figure 1: Receiver Characteristics Curve of ELISA in diagnosis of AML.
This table shows that there was statistical significant association (p<0.05) between IL17 plasma level results and blast cells in patients. IL17 ≥ 9.3 pg/mL was associated with high Blast count in peripheral blood and high Blast percentage in bone marrow (Table 3).
Table 3: Association between ELISA (IL17 plasma level) results and other laboratory characteristics in studied groups.
| Variables | IL17 plasma level | MW | P | |
|---|---|---|---|---|
| <9.3 pg/ml | ≥ 9.3 pg/ml | |||
| TLC (x103/ul): Median IQ-Range |
23.1 16.2–38.0 |
57.0 20.0–83.2 |
28.0 | 0.2 |
| Blast in peripheral blood (x103/ul): Median IQ-Range |
7.0 4.5–14.0 |
21.0 10.9–61.7 |
16.0 | 0.04 S |
| Blast in peripheral blood (%): Median IQ-Range |
31.0 20.0–53.0 |
78.5 28.5–87.3 |
26.0 | 0.2 |
| Blast in bone marrow (%): Median IQ-Range |
36.0 25.0–59.0 |
75.5 47.3–89.5 |
14.5 | 0.03 S |
This table shows that there was statistical significant difference (p<0.05) between patients and relatives in IL17F. Patients had significantly more GG genotype and G allele of IL17F than relatives, while relatives had significantly more AA genotype and A allele of IL17F than patients (Table 4).
Table 4: Genetic results in patients and relatives.
| PCR | Patients (n=27) | Relatives (n=40) | χ2 | P | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | ||||
| IL23R | Genotypes: GG AG AA UD |
8 19 0 0 |
29.6 70.4 0.0 0.0 |
8 31 0 1 |
20.0 77.5 0.0 2.5 |
1.8 | 0.2 |
| G allele | 27 | 100 | 39 | 97.5 | 0.01 | 0.9 | |
| A allele | 19 | 70.4 | 31 | 77.5 | 0.1 | 0.7 | |
| IL17A | Genotypes: GG AG AA UD |
5 17 3 2 |
18.5 63.0 11.1 7.4 |
13 25 0 2 |
32.5 62.5 0.0 5.0 |
2.8 | 0.09 |
| G allele | 22 | 81.5 | 38 | 95.0 | 1.9 | 0.2 | |
| A allele | 20 | 74.1 | 25 | 62.5 | 1.0 | 0.3 | |
| IL17F | Genotypes: GG AG AA |
7 7 13 |
25.9 25.9 48.2 |
3 12 25 |
7.5 30.0 62.5 |
3.8 | 0.05 S |
| G allele | 14 | 51.8 | 15 | 37.5 | 1.4 | 0.2 | |
| A allele | 20 | 74.1 | 37 | 92.5 | 4.3 | 0.03 (S) | |
This table shows that there was statistical significant difference (p<0.05) between patients and control group in IL17F. Patients had significantly more GG genotype and G allele of IL17F than control group, while control group had significantly more AA genotype and A allele of IL17F than patients (Table 5).
Table 5: Genetic results in patients and control group.
| PCR | Patients (n=27) | Control group (n=15) | χ2 | P | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | ||||
| IL23R | Genotypes: GG AG AA |
8 19 0 |
29.6 70.4 0.0 |
1 14 0 |
6.7 93.3 0.0 |
3.0 | 0.08 |
| G allele | 27 | 100 | 15 | 100 | NA | NA | |
| A allele | 19 | 70.4 | 14 | 93.3 | 1.8 | 0.2 | |
| IL17A | Genotypes: GG AG AA UD |
5 17 3 2 |
18.5 63.0 11.1 7.4 |
4 11 0 0 |
26.7 73.3 0.0 0.0 |
1.7 | 0.2 |
| G allele | 22 | 81.5 | 15 | 100 | fisher | 0.2 | |
| A allele | 20 | 74.1 | 11 | 73.3 | 0.01 | 0.9 | |
| IL17F | Genotypes: GG AG AA |
7 7 13 |
25.9 25.9 48.2 |
0 2 13 |
0.0 13.3 86.7 |
7.9 | <0.001 HS |
| G allele | 14 | 51.8 | 2 | 13.3 | fisher | 0.03 (S) | |
| A allele | 20 | 74.1 | 15 | 100 | fisher | 0.03 (S) | |
This table shows that there was statistical significant difference (p<0.05) between relatives and control group in IL17F. Control group had significantly more AA genotype of IL17F than relatives (Table 6).
Table 6: Genetic results in relatives and control group.
| PCR | Relatives (n=40) | Control group (n=15) | χ2 | P | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | ||||
| IL23R | Genotypes: GG AG AA UD |
8 31 0 1 |
20.0 77.5 0.0 2.5 |
1 14 0 0 |
6.7 93.3 0.0 0.0 |
1.6 | 0.2 |
| G allele | 39 | 97.5 | 15 | 100 | 0.01 | 0.9 | |
| A allele | 31 | 77.5 | 14 | 93.3 | 0.9 | 0.3 | |
| IL17A | Genotypes: GG AG AA UD |
13 25 0 2 |
32.5 62.5 0.0 5.0 |
4 11 0 0 |
26.7 73.3 0.0 0.0 |
0.1 | 0.7 |
| G allele | 38 | 95.0 | 15 | 100 | 0.01 | 0.9 | |
| A allele | 25 | 62.5 | 11 | 73.3 | 0.7 | 0.5 | |
| IL17F | Genotypes: GG AG AA |
3 12 25 |
7.5 30.0 62.5 |
0 2 13 |
0.0 13.3 86.7 |
4.1 | 0.04 S |
| G allele | 15 | 37.5 | 2 | 13.3 | Fisher | 0.1 | |
| A allele | 37 | 92.5 | 15 | 100 | Fisher | 0.5 | |
This table shows statistical significant association between IL17 plasma level results by ELISA and genetic results in studied groups. There were significant increase in GG (homozygous) and G allele of IL17F associated with increase in IL17 plasma level ≥ 9.3 pg/mL (cut off point) and decrease in GG (homozygous) and G allele were associated with decrease in IL17 plasma level <9.3 pg/mL (cut off point) (Table 7).
Table 7: Association between IL17 plasma level and genetic results in studied groups.
| PCR | IL17 plasma level | χ2 | P | ||||
|---|---|---|---|---|---|---|---|
| ˂9.3 pg/ml (n=45) |
≥ 9.3 pg/ml (n=37) |
||||||
| No. | % | No. | % | ||||
| IL23R | Genotypes: GG AG AA UD |
2 42 0 1 |
4.4 93.3 0.0 2.3 |
15 22 0 0 |
40.5 59.5 0.0 0.0 |
14.2 | ˂0.001 HS |
| G allele | 44 | 97.8 | 37 | 100 | Fisher | 0.9 | |
| A allele | 42 | 93.3 | 22 | 59.5 | Fisher | 0.003 (S) | |
| IL17A | Genotypes: GG AG AA UD |
5 33 3 4 |
11.1 73.3 6.7 8.9 |
17 20 0 0 |
45.9 54.1 0.0 0.0 |
14.3 | ˂0.001 HS |
| G allele | 38 | 84.4 | 37 | 100 | Fisher | 0.01 (S) | |
| A allele | 36 | 80.0 | 20 | 54.1 | 6.3 | 0.01 (S) | |
| IL17F | Genotypes: GG AG AA |
2 9 34 |
4.4 20.0 75.6 |
8 12 17 |
21.6 32.4 46.0 |
8.0 | 0.004 S |
| G allele | 11 | 24.4 | 20 | 54.1 | 7.6 | 0.005 (S) | |
| A allele | 43 | 95.6 | 29 | 78.4 | 5.6 | 0.01 (S) | |
The same results were found regarding GG homozygousity and G allele of IL17-A and IL23-R polymorphism, significant increase in GG homozygousity and G allele were found to be associated with increase in IL17 plasma level ≥ 9.3 pg/mL (cut off point) and decrease in GG homozygousity and G allele were associated with decrease in IL17 plasma level <9.3 pg/mL (cut off point).
Concerning AA homozygousity and A allele in IL-17F, IL17-A and IL23-R polymorphisms, we found that decrease of AA homozygousity and A allele is associated with increase in IL17 plasma level ≥ 9.3 pg/mL (cut off point) and increase in AA homozygousity and A allele is associated with decrease in IL17 plasma level <9.3 pg/mL (cut off point). So IL17 plasma level is directly proportional to GG homozygousity of IL-17F, IL17-A and IL23-R polymorphisms and G allele and inversely proportional to AA homozygousity and A allele of IL-17F, IL17-A and IL23-R polymorphisms (Table 8).
Table 8: ELISA (IL17 plasma level) results in studied groups.
| IL17 plasma level | Patients (n=27) | Relatives (n=40) | Control group (n=15) | χ2 | P | |||
|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |||
| ˂9.3 pg/ml | 4 | 14.8 | 30 | 75.0 | 11 | 73.3 | 26.1 | ˂0.001 HS |
| ≥ 9.3 pg/ml | 23 | 85.2 | 10 | 25.0 | 4 | 26.7 | ||
This table shows that there was high statistical significant difference (p<0.001) between patients and other two groups in IL17 plasma level results by ELISA. However, there was no statistical significant difference between relatives and control group in IL17 plasma level results by ELISA.
In the present study, we wanted to find out whether polymorphic variants of the genes coding for IL-17A, IL-17F and receptor for IL- 23 (as hallmarks of Th17 cells) could be associated with susceptibility to acute myeloid leukemia as it is implicated in many cancers as ovarian, esophageal, prostate, gastric, colorectal, hepatocellular, uterine, breast, lung, bladder, infection and inflammation.
Our results showed statistical significant increase in IL-17F GG (homozygous) & G allele among patients than other two groups (relatives and control group). Relatives showed non-statistical significant increase in GG (homozygous) & G allele than control group.
In contrast to the prevalence of the GG (homozygous) & G allele of IL17F among patients than other two groups (relatives and control group), the AA genotype and A allele of IL17F was increased significantly in relatives and control more than patients. As well control group showed statistical significant increase in AA genotype and A allele of IL17F more than relatives. So IL-17F gene G single mutant and GG homozygous mutant were more likely to be associated with acute myeloid leukemia susceptibility.
Zhu et al. showed that IL-17F gene G single mutant and GG homozygous mutant were associated with AML susceptibility. They revealed that AML patients exhibited significantly higher frequency of IL-17F G variant and homozygous than that of healthy controls [38]. Also Wróbel et al. found thatIL-17F G variant and its homozygosity were significantly more frequently observed among patients than healthy individuals [5]. These findings were in agreements with our results [5].
Concerning IL17-A, patients had non-statistical significant increase in AA homozygous and A allele and non-statistical significant decrease in GG homozygous and G allele than other two groups (relatives and control group).
Regarding IL23R, patients had non-statistical significant increase in GG homozygous than other two groups (relatives and control). Whereas patients and control had the same results regarding G allele of IL23R and relatives showed non-statistical significant decrease in G allele of IL23R than other two groups (relatives and control group). While AA homozygous of IL23R was absent in all three groups and A allele of IL23R was non-statistically significant decrease among patients than other two groups (relatives and control).
So IL17-A and IL23-R polymorphisms were not associated with occurrence of acute myeloid leukemia, this was matching with Zhu et al. who showed that IL17-A and IL-23-R gene polymorphism showed no correlation with occurrence of acute myeloid leukemia [38].
Interestingly, Kawaguchi et al. observed the functional consequences of the IL-17F polymorphism and suggested that the IL-17 expression and activity may be suppressed in carriers of the rare G allele [39]. That was in disagreement with our study. On studying IL-17F polymorphism, increase in GG (homozygous) and G allele were found to be associated with increase in IL17 plasma level ≥ 9.3 pg/mL (cut off point) and decrease in GG (homozygous) and G allele were associated with decrease in IL17 plasma level <9.3 pg/mL (cut off point).
The same results were found regarding IL17-A and IL23-R polymorphism, increase in GG homozygousity and G allele were found to be associated with increase in IL17 plasma level ≥ 9.3 pg/ mL (cut off point) and decrease in GG homozygousity and G allele were associated with decrease in IL17 plasma level <9.3 pg/mL (cut off point).
Concerning AA homozygousity and A allele in IL-17F, IL17-A and IL23-R polymorphisms, we found that decrease of AA homozygousity and A allele is associated with increase in IL17 plasma level ≥ 9.3 pg/mL (cut off point) and increase in AA homozygousity and A allele is associated with decrease in IL17 plasma level <9.3 pg/mL (cut off point). So IL17 plasma level is directly proportional to GG homozygousity of IL-17F, IL17-A and IL23-R polymorphisms and G allele and inversely proportional to AA homozygousity and A allele of IL-17F, IL17-A and IL23-R polymorphisms.
Our findings were in disagreement with Wróbel et al. who observed that patients carrying G allelic variant would be characterized by lower levels of IL-17 expression and they found no significant differences in the IL-17 plasma levels among AML patients with different IL-17F genotypes [5]. Also Zhu et al. did not find the significant differences of plasma IL-17 level in patients with different IL-17F genotypes [38]. While in the present study we found statistical significant association between plasma IL-17 level and IL-17F, IL17-A and IL23-R polymorphisms.
The IL-17 protein plasma level detected by Zhu et al. in AML patients was 8.8 ± 7.19 pg/mL (1.37-19.10 pg/mL). They did not find correlation between plasma IL-17 level and IL-17F gene polymorphism 38. While here in this study median of IL-17 protein plasma level in AML patients was 9.3 pg/mL and its level ranged from (5.0 to 18.02 pg/mL) and there was statistical significant association between plasma IL-17 level and IL-17F, IL17-A and IL23-R polymorphisms. Wróbel et al. detected plasma IL-17 level in AML patients at diagnosis (range, mean ± SD: 1.37–19.10, 8.8 ± 7.19 pg/mL), they found no significant relationships with respect to the IL-17 plasma levels and the three polymorphisms studied [5].
We also found in the current study that there were statistical significant association (p<0.05) and high statistical significant (p<0.001) association between IL17 plasma level by ELISA and the three genetic polymorphisms in studied groups. Also there was high statistical significant difference (p<0.001) between patients and other two groups in IL17 plasma level by ELISA.
Regarding correlation between IL17 plasma level and blast cells in AML patients, our results revealed significant association between them. IL17 plasma level ≥ 9.3 pg/mL was associated with high Blast count in peripheral blood and high Blast percentage in bone marrow in AML patients.
IL-17F gene G single mutant and GG homozygous mutant were associated with acute myeloid leukemia susceptibility in Egyptian population while IL17-A and IL23-R polymorphisms were not associated with susceptibility to the disease. Also there were statistical significant association (p<0.05) and high statistical significant (p<0.001) association between ELISA (IL17 plasma level) results and the three genetic polymorphisms in studied groups. There was high statistical significant difference (p<0.001) between patients and other two groups in IL17 plasma level by ELISA.
Citation: Mahfouz KA, Abo-Nar AA, Bendary SM (2018) Role of Interleukin17F (IL17F) Gene Polymorphism in Susceptibility to Acute Myeloid Leukemia. J Leuk 7:252. doi: 10.35248/2329-6917.19.7.252
Received: 21-Nov-2018 Accepted: 13-Jan-2019 Published: 30-Jan-2019 , DOI: 10.35248/2329-6917.19.7.252
Copyright: © 2019 Mahfouz KA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.