Research Article - (2016) Volume 2, Issue 1
Screening of Strong 1-Aminocyclopropane-1-Carboxylate Deaminase Producing Bacteria for Improving the Salinity Tolerance of Cowpea
*Corresponding Author: Nguyen Huy Thuan, Institute of Research and Development, Duy Tan University, K7/25 Quang Trung, Danang, Vietnam, Tel: +84-967-258-226, Fax: +84-5113-650-443 Email:
Abstract
Aim: To isolate rhizobacteria containing 1-aminocyclopropane-1-carboxylate (ACC) deaminase and evaluate the ability of selected bacteria for improving the growth of cowpea seedlings under salt stress conditions.
Methods: This study isolates salt-tolerant rhizobacteria which have strong ability to produce ACC deaminase and the phytohormone indol-3-acetic acid (IAA). Inoculation experiments with selected bacteria strains were used to verify the plant growth promoting activity of bacteria under salt stress conditions.
Result: Two isolates belong to Enterobacter cloacae and one isolate belongs to Pseudomonas sp. have been identified. Those rhizobacteria were found to be highly salt-tolerant at salinity level up to 10% NaCl. The selected bacterial strains were also capable to produce and secrete large amounts of ACC deaminase and the phytohormone IAA into the growth medium. Cowpea plants inoculated with ST3 strain revealed a significant increase in shoot length and shoot fresh weight over uninoculated control at the salinity level of 1.5% NaCl.
Conclusion: Three rhizobacterial strains belonging to the genera Enterobacter and Pseudomonas have been isolated. All three bacterial strains were identified as moderate halophiles and they can produce high levels of ACC deaminase and IAA. The strain Pseudomonas sp. ST3 showed the possible ability to promote the growth of cowpea under salt stress conditions.
Keywords: ACC deaminase; Cowpea, Enterobacter; Pseudomonas; Salinity tolerance
Introduction
Soil salinization from the agricultural standpoint is one of the most urgent problems in many areas worldwide, especially in agricultural countries like Vietnam. Salinization can be caused by saltwater invasion, long and severe drought, excessive use of chemical fertilizers. As a result, it can impact agricultural production, water quality, ecological health of streams and biodiversity. According to Qadir et al. [1], every day for the last two decades, about 2000 hectares of irrigated land in arid and semi-arid areas across 75 countries have been degraded by salt.
When exposed to salt stress conditions, plant tissues synthesize ethylene from its immediate precusor 1-aminocyclopropane-1- carboxylate (ACC). High levels of ethylene in plant tissues could inhibit the growth of root and shoot [2-4] as well as suppress leaf expansion [5]. Recently, researchers have found that several ACC deaminase producing bacteria can promote the growth of plants under salt stress conditions [6,7]. Such plant growth promoting bacteria (PGPR) belong to the genera such as Alcaligenes, Variovorax, Rhodococcus, Ochrobactrum and Bacillus [8,9]. Mechanism for this ability of PGPR is that enzyme ACC deaminase can hydrolyze ACC into amonium and α-ketobutyrate, and as a result, the ACC level in plant tissues is decreased [10,11].
Beside the ability to produce ACC deaminase enzyme, some PGPR belonging to the genera Alcaligenes, Bacillus and Ochrobactrum sp. can also synthesize the phytohormone Indole-3-acetic acid (IAA) [8]. Previous studies have shown that inoculation with PGPR producing both ACC deaminase and IAA can enhance the salt tolerance and consequently improving the growth of plants under salt stress conditions. Such PGPR have shown the ability to improve the growth of soybean and maize under salt stress in the field conditions, however, these effects are still limited [12,13].
To date, little information is available on the isolation, identification and application of rhizobacteria for improving crops growing under salt stress conditions in Vietnam. Therefore, the present study aims to isolate the strong ACC deaminase and IAA producing rhizobacteria from salt-affected area of Danang, Vietnam. Furthermore, the abilities of selected rhizobacteria to promote the growth of cowpea under saltstress conditions were also investigated.
Materials and Methods
Isolation of ACC deaminase producing rhizobacteria
Soil samples were collected from the rhizosphere of water spinach (Ipomoea aquatica L.) in the depth of 0-10 cm at Son Tra peninsula, Danang, Vietnam (16°10′ N, 108°22′ E). The rhizobacteria in soil samples were isolated by the method of Penrose and Glick [11]. The bacterial isolates observed by using this specific isolation method are known to have the ability to use ACC as the sole nitrogen source. These selected isolates were stored at –80°C for further experiments.
Identification of bacterial strains
In brief, the purified genomic DNA from 3 isolates was used as the templates for the amplification of conserved region of the 16S rRNA gene with the following universal primers: 27 F 5′- AGAGTTTGATCCTGGCTCAG-3′; 1492 R 5′- GGTTACCTTGTTACGAC TT-3′. The PCR products were then checked on 0.8% agarose gel. The DNA band of the 16S rRNA gene (approximately 1500 bp) was cut and purified by GeneJET Gel Extraction Kit (Thermo Scientific). The purified PCR products were then sent to Eurofins (Germany) for sequencing. The full-length of the 16S rRNA gene sequence (1500 nucleotides) was determined by direct sequencing of PCR-amplified 16S rDNA. The nucleotide sequences obtained were compared to the references of the 16S rRNA gene sequences retreived from GenBank database by using BLAST (NCBI, USA).
Analysis of the ACC deaminase activity
Selected bacterial strains were grown in tryptic soybean broth (TSB) medium at 30°C, 200 rpm for 24 h. Bacterial cells were then transferred to the Dworkin and Foster salt minimal medium (DF) containing 3 mM ACC as the sole nitrogen source and grown at 30°C, 200 rpm for 48 h to induce the production of ACC deaminase. Culture supernatant was collected at 12, 24, 36 and 48 h. The ACC deaminase activity was determined by measuring the production of α- ketobutyrate generated by the cleavage of ACC according to the protocol described by Penrose and Glick [11]. The amount of α- ketobutyrate produced by this reaction was measured by comparing the absorbance at 540 nm of the sample to a standard curve of α- ketobutyrate (Sigma-Aldrich Co, USA) ranging between 0.1 and 1.0 μM. The ACC deaminase activity was expressed as the amount of α- ketobutyrate produced per mg of protein per hour.
Analysis of the bacterial salt tolerance
Each bacterial strain was grown in 6 flasks containing 50 mL Nutrient Broth (NB) medium supplemented with the following concentrations of NaCl: 0%, 2%, 4%, 6%, 8% and 10% (w/v). The flasks were incubated at 30°C, 200 rpm for 48 h and the absorbance at 600 nm of each sample was measured afterwards. The standard deviation based on 3 independent cultivations.
Analysis of the IAA production
In order to analyze the IAA production, each bacterial strain was grown in DF salt minimal medium containing 3 mM ACC supplemented with 2 mg/mL L-tryptophan (Sigma-Aldrich Co, USA) at 30°C, 200 rpm. A 2 mL of cell culture supernatant was collected after 12, 24, 36 and 48 h of cultivation by centrifugation. The IAA concentration in the cell culture supernatant was measured using the colorimetric technique as described by Gordon and Weber [14]. In brief, the cell culture supernatant containing IAA was mixed with Salkowski reagent (2:1) and incubated at room temperature for 30 minutes in dark. The IAA concentration was measured by comparing the absorbance at 530 nm of a sample to the standard curve of IAA (Sigma-Aldrich Co, USA) ranging between 5 and 100 μg/mL. The standard deviation based on 3 independent cultivations.
Effect of bacteria on plant growth under salt stress conditions
In this experiment, cowpea (Vigna unguiculata) seeds were surfacedisinfected by immersion in 70% (v/v) ethanol plus 0.3% (v/v) Tween 80 for 5 minutes and later by solution containing 3% (w/v) HCl and 0.3% (v/v) Tween 80 for 20 minutes. Seeds were then washed three times with sterile distilled water. For germination, sterilled cowpea seeds were soaked in warm water (60°C) for 12 h and then covered with a clean cloth for 12 h. Soil for planting was sterilized at 121°C, 1.2 atm for 30 minutes in an autoclave and then transferred to planting pots (20 cm diameter x 15 cm height).
For each crop, four sets of pots with three replicates were prepared: control (applied water only), NaCl (applied 1.5% NaCl solution), T3 (applied ST3 cells solution), T3 + NaCl (applied ST3 cells solution and 1.5% NaCl solution). For each pot, 20 germinated cowpea seeds were transplanted at the same depth (approx. 2 cm below the surface). The plants were grown under natural light and temperature (14 h photoperiod, 25-35°C). Salility was imparted to the plant by adding 1.5% (w/v) NaCl solution to the rhizosphere of the plants at day 7 after planting, three times a week. The bacterial strain ST3 was grown in DF salt minimal medium (108 CFU/mL) and centrifuged to collect the cell pellets. Bacterial cell pellets were then resuspended in 200 mL sterile distilled water and applied to the plant rhizosphere at day 7 after planting, once a week. Seedlings were harvested on day 30 for measuring the shoot length and shoot fresh weight.
Data regarding the effect of bacteria on shoot length and shoot fresh weight of cowpea were statistically analyzed by one-way analysis of variance (ANOVA), followed by Duncan’s multiple range test (DMRT) at p ≤ 0.05. Statistical analysis was performed using the R software for Windows (version 3.2.2).
Results
Screening of salt tolerant PGPR producing ACC deaminase
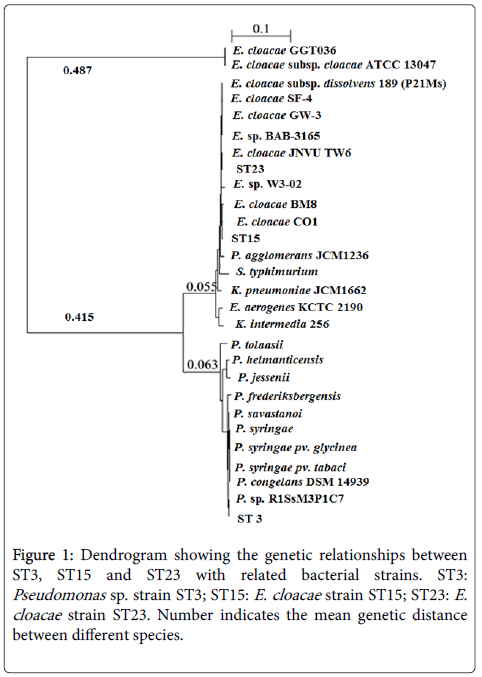
In the screening step, among twenty-five isolates selected from soil samples, only three isolates designated ST3, ST15 and ST23 were able to use ACC as the sole nitrogen source. The genomic DNA samples from those isolates were then isolated and used as templates for the amplification and subsequent sequencing of the 16S rRNA genes. The 16S rRNA gene sequence similarity values for isolates ST3, ST15, ST23, and related strains based on the partial sequence comparison were analyzed (data not shown). It was found that ST15 and ST23 belong to the Enterobacter genus, while ST3 showed highly close genetic relationship with Pseudomonas. In particular, ST15 and ST23 showed high 16S rRNA gene similarity of more than 99% with Enterobacter cloacae strain JNVU TW6 (accession number: 342359686) or Enterobacter cloacae strain CO1 (accession number: 572486716). In a similar manner, ST3 shared high identity value with Pseudomonas sp. R1SsM3P1C7 (accession number: 530252117). Therefore, the selected bacteria were named as Pseudomonas sp. ST3, E. cloacae ST15 and E. cloacae ST23, respectively. The obtained 16S rRNA gene sequences were then deposited into the GeneBank Nucleotide Sequence Database (NCBI) under accession numbers KU049661, KU049659 and KU049660 (not yet released) for strains ST3, ST15 and ST23, respectively. The phylogenetic tree between ST3, ST23, ST15 and related taxa (Figure 1).
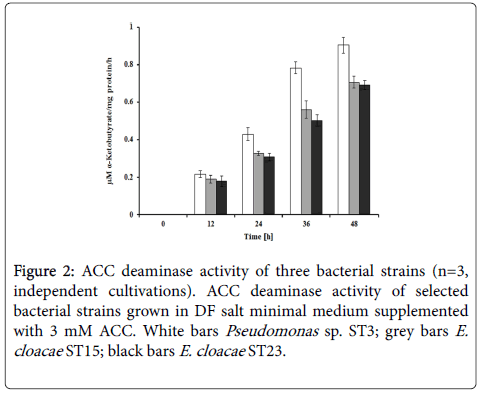
Data of ACC deaminase assay revealed that all three selected strains showed the ACC deaminase activities ranging between 0.7 and 0.9 μM α-ketobutyrate/mg/h after 48 h of cultivation (Figure 2). Among three strains, ST3 showed the highest ACC deaminase activity of about 0.9 μM α-ketobutyrate/mg/h.
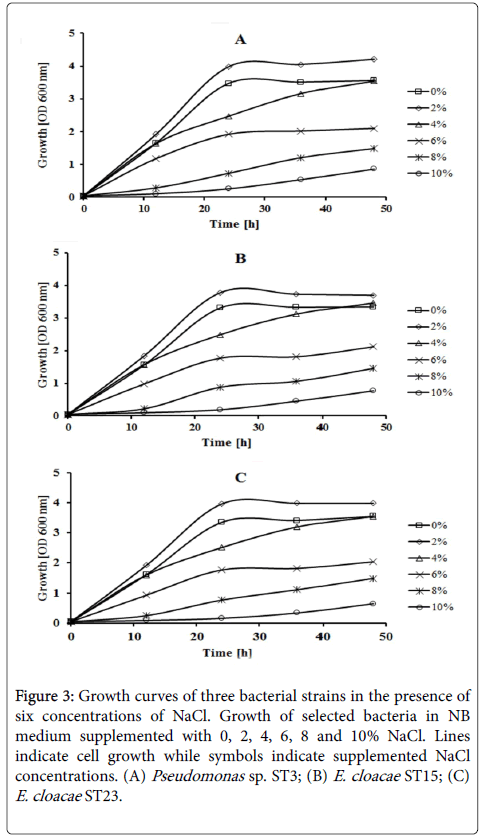
Screening of bacterial strains for high salinity tolerance showed that all three bacterial strains were able to grow well at the salinity level ranging between 0-6% (w/v) NaCl. The growth rates were highest for all bacterial strains in the medium supplemented with 2% (w/v) NaCl (Figure 3). Although the growth rate of the selected bacterial strains was decreased when concentrations of NaCl increased, those strains exhibited the ability to grow at high NaCl concentrations up to 10% (w/v).
Figure 3: Growth curves of three bacterial strains in the presence of six concentrations of NaCl. Growth of selected bacteria in NB medium supplemented with 0, 2, 4, 6, 8 and 10% NaCl. Lines indicate cell growth while symbols indicate supplemented NaCl concentrations. (A) Pseudomonas sp. ST3; (B) E. cloacae ST15; (C)E. cloacae ST23.
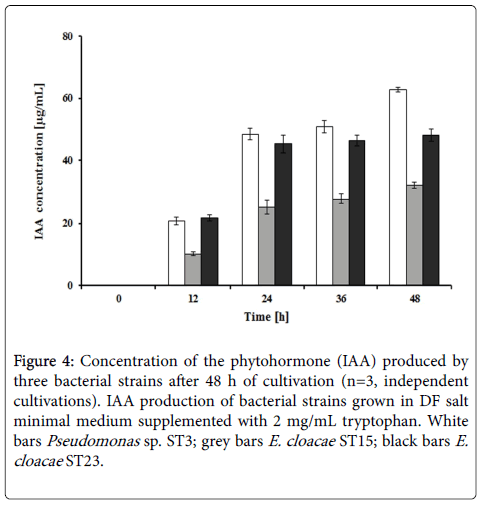
In this study, the plant growth promoting characteristic of bacterial strains based on the ability to produce phytohormone IAA was also investigated. When grown in DF salt minimal medium supplemented with 2 mg/mL tryptophan all three bacterial strains started to produce IAA after 12 h of cultivation and the amount of IAA reached the maximum values at 48 h (Figure 4). This result also demonstrated that the strain ST3 produced higher amount of IAA when compared to the two strains ST15 and ST23. Analysis showed that the highest positive correlation value (Pearson Correlation) between the IAA and ACC deaminase production was observed for strain ST3 (r=0.901, p ≤ 0.01). Strains ST15 and ST23 had lower r values of 0.894 and 0.777 (p ≤ 0.01), respectively.
Figure 4: Concentration of the phytohormone (IAA) produced by three bacterial strains after 48 h of cultivation (n=3, independent cultivations). IAA production of bacterial strains grown in DF salt minimal medium supplemented with 2 mg/mL tryptophan. White bars Pseudomonas sp. ST3; grey bars E. cloacae ST15; black bars E.cloacae ST23.
Effect of selected bacterial strains on growth of cowpea under salt stress conditions
Among three selected bacterial strains, ST3 exhibited higher potential for improving growth of plant under salt stress conditions. Therefore, ST3 strain was selected for the inoculation experiment. In this experiment, strain ST3 was applied as an aqueous suspension to the rhizosphere of cowpea one week after planting. As shown in table 1, the application of ST3 cells to the pot T3 resulted in significantly increased of the shoot length and shoot fresh weight of cowpea seedlings (31.9% and 43.3% more than the uninoculated control, respectively). It was not surprised that the lowest values of shoot length and shoot fresh weight were observed for the pot NaCl due to the salt stress. However, the shoot length values increased up to 21.7% when strain ST3 was applied to the pot T3+NaCl as compared to the pot NaCl.
| Treatment | Shoot length (cm) | Shoot fresh weight (g) |
|---|---|---|
| Control (fresh water only) | 14.1ab ± 0.17 | 18.7ab ± 0.08 |
| NaCl (with 1.5% NaCl solution) | 11.5b ± 0.14 | 11.7b ± 0.09 |
| T3 (with ST3 cells) | 18.6a ± 0.25 | 26.8a ± 0.07 |
| T3+NaCl (1.5% NaCl with ST3 cells) | 14ab ± 0.1 | 22.7a ± 0.05 |
Note: Means ± SEM (Standard error of means) of triplicates with 20 seedlings for each treatment (n=60). Mean values sharing different superscript letters in column are significantly different according to Duncan’s multiple range test (p ≤ 0.05).
Table 1: Shoot length and shoot fresh weight of cowpea after 30 days of treatment.
Although, the shoot length values between the T3+NaCl and the control pots are not significantly different (p ≤ 0.05), the application of the ST3 strain to the pot T3+NaCl resulted in significantly higher shoot fresh weight (21.4% more than the control pot) (p ≤ 0.05). The different appearances of cowpea seedlings in figure 5 had also supported those results.
Discussion
This study indicates the effectiveness of ACC deaminase producing rhizobacteria for improving the salt tolerance of cowpea plants. In this study, soil bacteria have been isolated from rhizosphere of water spinach (I. aquatica L.). As reported by Yousif et al. [15], the growth of water spinach is markedly reduced under saline conditions. However, the existence of water spinach in salt-affected area suggesting that the rhizobacteria colonized its roots may contribute to the salt tolerance of this plant. According to the salt requirement, bacteria may be classified as: non-halophiles grow at NaCl concentration ranging between 0-2%; slight halophiles grow at 2-3% NaCl; moderate halophiles grow at 5-10% NaCl; and extremely halophiles grow at NaCl concentration greater than 10% [16]. Data of this study revealed that all three selected bacteria were able to proliferate at 10% NaCl and thus they can be classified as moderate halophiles. As reported by Bal et al. [8], the other ACC deaminase producing bacteria belonging to the genus Bacillus and Ochrobactrum can only grow in maximum NaCl concentration of about 6%. Bacterial strains belonging to the genus Pseudomonas and Enterobacter have been previously reported as salt tolerant organisms [17,18]. Paul and Nair reported that Pseudomonas fluorescens MSP-393 can produce osmolytes and salt-stress induced protein that overcome the negative effect of salt [19].
Data of this study demonstrated that all three selected strains were able to produce both ACC deaminase and IAA at significantly high concentrations. The ability to produce both components have been found in several bacterial species belonging to the genera such as Bukholderia, Alcaligenes, Ochrobactrum and Bacillus [20,21]. The ACC deaminase producing P. fluorescens spp. have been reported to produce IAA as well [6,22]. However, the capacity to produce IAA of those bacterial strains is significantly lower than our selected bacterial strains (Table 2).
| S. No | Bacterial strains | ACC deaminase activity (μM α-Ketobutyrate/mg/h) | IAA (μg/mL) | References |
|---|---|---|---|---|
| 1 | B. pumilusSB1-ACC3 | 1.46 | 45.91 | [8] |
| 2 | B. licheniformisB2r | 0.86 | N/A | [23] |
| 3 | Enterobacter sp. CS1 | 0.17 | N/A | [7] |
| 4 | P. fluorescensP4 | + | 8.93 | [6] |
| 5 | P. fluorescensACC50 | 0.30 | 15.3 | [24] |
| 6 | Pseudomonas sp. PDMZnCd2003 | N/A | 11.15 | [25] |
| 7 | Pseudomonas sp. Crb56 | + | 18.5 | [22] |
| 8 | Pseudomonas sp. ST3 | 0.90 | 62.7 | This study |
| +: Positive; NA: Not Available | ||||
Table 2: The ACC deaminase activity and IAA production of different bacterial species.
Although the ability to produce ACC deaminase and IAA are particularly important for bacteria to improve the growth of plant under salt stress conditions, little information is available for the correlation between these two factors in relation to plant growth promotion. In this study, the ST3 strain showed the highest positive correlation between the ACC deaminase and IAA production ability among all selected strains. According to Glick [26,27], bacterial IAA together with endogenous plant-synthesized IAA can induce the transcription of ACC synthase and thus increase the level of ethylene in plant. However, as plant ethylene levels increase, ethylene will inhibit the IAA signal transduction and thereby limiting the extent that IAA can induce the ACC synthase transcription [28,29]. Furthermore, the large portion of synthesized ACC is degraded by the bacterial ACC deaminase. Therefore, the net result of the interaction between IAA and ACC deaminase is that the ethylene level in plant is reduced and IAA still stimulates the growth of plant.
The positive effects of the efficient salt-tolerant bacteria containing ACC deaminase for the growth, yield and disease suppression of different plants under salt stress conditions have been reported [18,26]. The role of Pseudomonas spp. for promoting salt resistance of rice and groundnut have also reported [24,30]. However, the effects of those bacterial strains for enhancing plant growth under salt stress conditions are still limited. Our data suggest that the Pseudomonas sp. strain ST3 is an effective rhizobacteria for enhancing the growth of cowpea under salt stress conditions. This could be due to the particularly high ACC deaminase and IAA production abilities of ST3 strain. Pseudomonas species have been reported to solubilize phosphate sources [31] and reduce the development of root-rot disease and enhance the yield of Phaseolus vulgaris L. [32]. Therefore, ST3 could be a promising candidate for potential biofertilizers in saline fields. Further work is needed to evaluate the effectiveness of ST3 strain for improving the plant growth under salt-affected field conditions.
Conclusion
Three rhizobacterial strains belonging to the genera Enterobacter and Pseudomonas have been isolated in this study. All three bacterial strains were salt resistance and they can produce high levels of ACC deaminase and IAA. The strain Pseudomonas sp. ST3 showed the possible ability to promote the growth of cowpea under salt stress conditions.
Acknowledgements
We thank Institute of Research and Development, Duy Tan University for providing facilities for this research. We are grateful to collaborators and students who have contributed to this work.
References
- Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, et al. (2014) Economics of salt-induced land degradation and restoration. Natural Resources Forum 38: 282-295.
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell DevBiol 16: 1-18.
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221-233.
- Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant 118: 10-15.
- Peterson TA, Reinsel MD, Krizek DT (1991) Tomato (Lycopersiconesculentum Mill., cv. ‘Better Bush’) plant response to root restriction. II. Root respiration and ethylene generation. J Exp Bot 42: 1241-1249.
- Akhgar AR, Arzanlou M, Bakker PAHM, Hamidpour M (2014) Characterization of 1-aminocyclopropane-1-carboxylate (ACC) deaminase-containing Pseudomonas spp. in the rhizosphere of salt-stressed canola. Pedosphere 24: 461-468.
- Huang H, Huang M, Gan G, Liu X, Wang J, et al. (2013) Isolation and characterization of 1-aminocyclopropane-1-carboxylate (ACC) deaminase-containing plant growth-promoting rhizobacteria from carnation soil and roots. Afr J Microbiol Res 7: 5664-5668.
- Bal HB, Nayak L, Das S, Adhya TK (2013) Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant and Soil 366: 93-105.
- Belimov AA, Hontzeas N, Safronova VI, Demchinskaya SV, Piluzza G, et al. (2005) Cadmium-tolerant plant growth-promoting rhizobacteria associated with the roots of Indian mustard (Brassica junceaL. Czern). Soil BiolBiochem 37: 241-250.
- Glick BR (1995) The enhancement of plant growth by free living bacteria. Can J Microbiol 41: 109-117.
- Glick BR, Penrose DM, Li J (1998) A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria J TheorBiol 190: 63-68.
- Husen E, Wahyudi AT, Suwanto A, Giyanto (2011) Soybean response to 1-aminocyclopropane-1-carboxylate deaminase-producing Pseudomonas under field soil condition. Am J AgriBiolSci 6: 273-278.
- Zafar-ul-Hye M, Farooq HM, Zahir ZA, Hussain M, Hussain A (2014) Application of ACC-deaminase containing rhizobacteria with fertilizer improves maize production under drought and salinity stress. Inter J AgriBiol 16: 591-596.
- Gordon SA, Weber RP (1951) Colorimetric estimation of indoleacetic acid. Plant Physiol 26: 192-195.
- Yousif BS, Nguyen NT, Fukuda Y, Hakata H, Okamoto Y, et al. (2010) Effect of salinity on growth, mineral composition, photosynthesis and water relations of two vegetable crops; New Zealand spinach (Tetragoniatetragonioides) and water spinach (Ipomoea aquatica). Inter J AgriBiol 12: 211-216.
- Agwu O, Oluwagunke T (2014) Halotolerance of heterotrophic bacteria isolated from tropical coastal waters. Inter J Sci 16: 224-231.
- Egamberdieva D, Botir H, Abeer H, Abd-Allah EF (2014) Characterization of salt tolerant Enterobacter hormaechei strain associated with tomato root grown in arid saline soil. J Pure ApplMicrobiol 8: 4231-4239.
- Wang C, Knill E, Glick BR, Defago D (2000) Effect of transfering 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase gene into Pseudomonas fluorescens strain CHAO and its gacA derivative CHA96 on their growth-promoting and disease-suppresive capacities. Can J Microbiol 46: 898-907.
- Paul D, Nair S (2008) Stress adaptations in a Plant Growth Promoting Rhizobacterium (PGPR) with increasing salinity in the coastal agricultural soils. J Basic Microbiol 48: 378-384.
- Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541-556.
- Onofre-Lemus J, Hernández-Lucas I, Girard L, Caballero-Mellado J (2009) ACC (1-aminocyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Appl Environ Microbiol 75: 6581-6590.
- Husen E, Wahyudi AT, Suwanto A, Saraswati R (2009) Soybean seedling root growth promotion by 1-aminocyclopropane-1-carboxylate deaminase-producing Pseudomonas. Indo J AgriSci 10: 19-25.
- Chookietwattana K, Maneewan K (2012) Selection of salt-tolerant bacteria containing ACC deaminase for promotion of tomato growth under salinity stress. Soil Environ 31: 30-36.
- Shaharoona B, Jamro GM, Zahir ZA, Arshad M, Memon KS (2007) Effectiveness of various Pseudomonas spp. and Burkholderiacaryophilli containing ACC-deaminase for improving growth and yield of wheat (TriticumaestivumL.). J MicrobiolBiotechnol 17: 1300-1307.
- Nakbanpote W, Panitlurtumpai N, Sangdee A, Sakulpone N, Sirisom P, et al. (2014) Salt-tolerant and plant growth-promoting bacteria isolated from Zn/Cd contaminated soil: identification and effect on rice under saline conditions. J Plant Interact 9: 379-387.
- Shahzad SM, Khalid A, Arshad M, Kalil-ur-Rehman (2010) Screening rhizobacteria containing ACC-deaminase for growth promotion of chickpea seedlings under axenic conditions. Soil Environ 29: 38-46.
- Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169: 30-39.
- Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, et al. (2007) Promotion of plant growth by ACC deaminase. Crit Rev Plant Sci 26: 227-242.
- Pierik R, Tholen D, Poorter H, Visser EJ, Voesenek LA (2006) The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci 11: 176-183.
- Saravanakumar D, Samiyappan R (2007) ACC deaminase from Pseudomonas fluorescens mediated saline resistance in groundnut (Arachis hypogea) plants. J ApplMicrobiol 102: 1283-1292.
- Bhardwaj D, Ansari MW, Sahoo RK, Tuteja N (2014) Biofertilizers function as key player in sustainable agriculture by improving soil fertility, plant tolerance and crop productivity. Microb Cell Fact 13: 66.
- Neeraj KS (2011) Organic amendments to soil inoculated Arbuscularmycorrhizal fungi and Pseudomonas fluorescens treatments reduce the development of root-rot disease and enhance the yield of Phaseolus vulgaris L. Eur J Soil Biol 47: 288-295.
Copyright: © 2016 Thuan NH, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.