Journal of Hepatology and Gastrointestinal disorders
Open Access
ISSN: 2475-3181
ISSN: 2475-3181
Research Article - (2022)Volume 8, Issue 4
Prolonged administration of antiviral therapy for chronic hepatitis B may result in the development of hepatitis B viral mutants. To gain insight into the mechanism involved in the sequence variability of Hepatitis B Virus (HBV) surface antigen gene (S gene) among responders and non-responders to antiviral therapy, baseline characteristics of the patients and sequences within the S region were investigated in pre-treatment serum samples of responders and post-treatment serum samples of non-responders. The data was collected from 15 individuals with chronic hepatitis B from Khyber Pakhtunkhwa (KPK) province, Pakistan. The antiviral response was independent of viral genotypes, and nonresponse to antiviral therapy was associated with a complex variability of the viral mutants. The sequence analysis of the S gene among responders and non-responders patients of pre and post-treatment with antiviral therapy showed variability in DNA sequence marked as Pakistani isolates make a distinct cluster in the phylogenetic tree. The S gene of HBV isolates from KPK province shows some similarities with isolates of other countries. No significant variations of nucleotides in the S gene of HBV was found among the responders and non-responders receiving antiviral therapy indicating that S gene may not be important with respect to treatment outcome. It illustrates that antigenicity of other various HBV proteins can be targeted in order to design more effective vaccines against the local strains.
Hepatitis B Virus (HBV); Surface mutations; Surface gene; Sequence analysis
HBV: Hepatitis B Virus; KPK: Khyber Pakhtunkhwa; S gene: Surface Antigen Gene; HBeAg: Hepatitis B e Antigen; HbsAg: Hepatitis B Surface Antigen; RT: Reverse Transcriptase; ICT: Immuno Chromatography Technique
Hepatitis B is a serious public health problem worldwide. Initially it causes acute liver diseases that might also lead to chronic stages including liver cirrhosis and hepatocellular carcinoma [1]. Studies showed that about two billion of people are infected worldwide from the time of its discovery that is expectedly causing about 600,000 deaths annually [2,3]. Hepatitis B is caused by Hepatitis B Virus (HBV), which falls in the Hepadna virus family and is believed to the only human demonstrative of this family. HBV is categorized into eight genotypes, i-e A to H, which is based on the intra-group nucleotide deviation in S genome and whole genome sequences up to 4.2% and >8%,respectively which comprises of about 3200 base pairs [3-5]. HBV has a rounded and partly double stranded DNA genome with 3.2 kb in size which contains four overlying open reading frames [6]. The four known genes of HBV encoded by the genome are called C, P, S and X. Gene S codes for the surface antigen (HBsAg) which has been defined as the molecular basis for the serologic heterogeneity [7]. Moreover, on the basis of limited sequencing contained by the S gene, the HBV genotyping is viable, with results reliable with those established on the sequencing of complete genomes [8-10].
Mutation in the S region of genome of the variants of hepatitis B virus (HBV) occurred and emerged reported that the genome of Hepatitis B virus (HBV) indicate a high rate of mutations, resulting in a variety of changes in the amino acid, which can change the conformation of viral proteins, as a result antibody binding is reduced or the secretion of surface antigen (HBsAg) is diminished. Few types of pre-S mutants in patients arising naturally in the hepatic tissue or serum infection have been noticed. Whereas, in molecular based assays and immunologic sensitivity, a challenge is represented by the genetic variability of hepatitis B virus (HBV) [11]. Along with, a structurally modified nucleotide/nucleoside analogue exerts potent inhibitory effect on HBV polymerase activities [10]. During long-term therapy, mutations happened not merely in the polymerase gene but also in the S gene, produced increase of two types of surface protein mutants. It was also recognized that 5%-10% hepatitis B adult vaccines were non-and hypo-responders and possibly was not effectively protected against Hepatitis B Virus (HBV) infection affecting the duration and intensity of protective humoral immune response to the hepatitis B vaccine.
A study on vaccinations has been made but certain limitation suggests further improvement in term of long term effects. In addition to regular an evolutionary variation that participates in genetic changes, vaccination or antiviral therapy also contribute in the mutations process. Okamoto et al. stated that a variant of hepatitis B virus (HBV) having a specific mutation within the S gene has been found to infect vaccines [12,13]. Yamamoto et al. reported that HBV escape mutants with mutations in the S gene affecting the expression of group-specific determinants would survive in some carriers after they seroconvert to antibody against surface antigen [13]. Carriers with HBV escape mutants may transmit HBV either by donation of blood units without detectable surface antigen or through community-acquired infection, which would hardly be prevented by current hepatitis B immunoglobulin or vaccines. The lack of identical sites in most of its coding sequence and also the frequent change in the amino acid of its pre-core and S gene region give an additional property towards successful dogging the immune system [14]. Based on the perspective, the central hypothesis of this study is to investigate the sequence variability of S gene of HBV among the non-responders of antiviral therapy by amplification and sequencing of the S gene of HBV by qualitative PCR. Moreover, the genetic linkage was carried out phylogenetic analysis using different bioinformatics tools.
Hepatitis B serology analysis
All patients were screened for HBsAg using rapid test strip (ACON Laboratories Inc. San Diego. CA, USA). The confirmation of all HBsAg positive were made using 3rd generation enzyme linked immunosorbent assay (Bio-Rad Berkeley, California) according to manufacturer’s instructions. Initial screening was conducted through Immuno Chromatography Technique (ICT) (BD USA) strips.
All the patients were further confirmed by the presence of HBV through ELIZA (Bio-Rad Berkeley, California). Serum was separated for serology tests by centrifuging whole blood 5000 rpm for 4 min using micro centrifuge (Sigma D-37520, Germany). The serum samples were then stored at -20ºC immediately.
Antiviral therapy analysis
Alpha interferon therapy treatment was done for 3 months. And then their serum was taken and PCR (T100™ Thermal Cycler BIO RAD USA) was performed, patients who were still positive and did not respond to the antiviral therapy (non-responder) and contained the virus in their blood was further investigated in the study. Whereas, patients whose PCR results came negative and cleared all the viruses from their blood and showed response to the antiviral therapy were classified as responder.
Amplification and documentation of HBV surface gene (S gene)
HBV DNA was extracted from both antiviral responder and nonresponder samples using commercial viral DNA extraction kit (Favorgen Biotech Corporation, Pingtung, Taiwan) according to manufacturer’s instructions. The extracted HBV DNA was stored at -20°C prior to DNA amplification for further use. The extracted stored HBV DNA was of responders and nonresponders were used for the amplification of S gene. The extracted DNA was amplified using S-gene specific primer i.e. Forward Primer 5/-GGTATGTTGCCCGTTTGTCCTCT-3/ and Reverse primer 5/-GGCACTAGTAAACTGAGCCA-3/. Nested PCR reaction was carried out using a total volume of 20 μL including 2 μL of DNA extract, 1 μL of forward primer, 1 μL of reverse primer, 4 μL of Magnesium chloride, 0.5 μL of Taq polymerase, 2 μL of Taq buffer, 2 μL of DNTPs and 7.5 μL of distilled water. Amplification was carried out in PCR (T100™ Thermal Cycler BIO RAD USA) using the following protocol: Initial denaturation at 95°C for 5 min, 30 cycles at 95°C for 30 seconds, 57°C for 30 seconds and 72°C for 30 seconds and final extension at 72°C for 10 minutes. The amplified PCR products were visualized using 2% agarose gel pre-stained with ethidium bromide. Visualization of amplified PCR products was made using (Uvitec Limited, Cambridge, UK) gel documentation system.
Phylogenetic analysis of S gene
The S gene product was separated from agarose gel and purified using gel purifying kit (Thermo Fisher Scientific, USA) according to the manufacturer's instructions. Sequencing was performed at Korea through Macrogen by using Big Dye Deoxy Terminator method. (Applied Biosystems) with the help of a private company (Worldwide Scientific Islamabad). Phylogenetic tree of the HBV S gene nucleotide sequences isolated from Responders and non-responders was constructed with the HBV Surface gene sequences retrieved from NCBI of various geographical regions of the world by using Mega 6.
Antiviral therapy analysis
Since Hepatitis B virus (HBV) infection is described by a high rate of development of chronic infection with either high-or lowtiter viremia [15], the results of which include cirrhosis or hepatocellular carcinoma [16]. Variability in the genome of HBV is involved as one of the mechanisms in viral existence. In fact, HBV duplicates its DNA genome via a reverse-transcription step [17], and the rate of spontaneous error of the reverse transcriptase (RT) proceeds to the occurrence of mutations which may collected or be selected during the course of infection. Eight genotypes (A to H) of hepatitis B virus (HBV) have been defined so far [18]. For the identification of sequence variability of HBV S gene among the 10 non-responder samples of antiviral therapy, 5 responder samples were also taken as control group. Initial screening was done through ICT and ELIZA followed by PCR. The study involved 150 HBV infected patients who were referred to several health care units of KP; qualitative detection of HBV DNA was done for finding of active infection. Among 150 patients, only 10 patients did not show response to the antiviral therapy and the rest showed response and cleared the Viral DNA. From Figure 1, 6.66% patients with active infection were detected while 93.33% patients were found to have no infection after taking antiviral therapy. About 5 (3.33%) PCR positive samples of responders were investigated. Each genotype appears to have a limited geographical distribution; consequently, it establishes an invaluable means in finding HBV molecular progression and the arrangement and methods in which HBV spreads [18]. This variety may be accountable for the selection of mutants that escape the immune response to the hepatitis B e-antigen (HBeAg; i.e., the pre core region mutants), the immune response to the hepatitis B surface antigen (HBsAg; i.e., the surface gene mutants), or the capsid T cell response (i.e., the core gene mutants) [15]. On the other hand, mutants may be more modified to definite host circumstances, such as immunodeficiency or antiviral therapy, and may outgrow wild type virus [19].
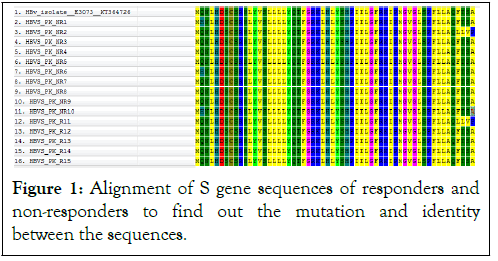
Figure 1: Alignment of S gene sequences of responders and non-responders to find out the mutation and identity between the sequences.
Identification of S gene
The basic purpose of current study was to identify and characterize the sequence variability of HBV S gene among the non-responders of antiviral therapy. For this reason we designed a study to find out HBV S gene sequences of responders and non-responders by qualitative PCR followed by sequencing among the actively infected patients in Figure 2. The samples selection was irrespective of the gender. The emergence and takeover of hepatitis B virus (HBV) variants carrying mutation(s) in the preS/S genomic region is a fairly frequent event that may occur spontaneously or may be the consequence of immune prophylaxis or antiviral treatments. Selection of preS/S mutants may have relevant pathobiological and clinical implications. The variation of the natural course of the infection and related disease is determined by the interaction between virus and host factors [19]. Several lines of evidence indicate that a certain number of HBV genetic variants, apparently provided with higher pathogenicity, may emerge during the course of the infection under endogenous (host immunity) and/or exogenous (immune prophylaxis and antiviral therapies) selection pressures [20]. In our study, 5 responder and 10 non-responder samples of HBV were sequenced for 230 bp fragment of surface gene, which make the envelop protein for HBV. After sequencing and bioinformatics analysis we find out that our isolates of HBV formed a distinct cluster in the phylogenetic tree. This showed that Pakistani isolates of HBV have distinct sequences as compared to other countries and showing a great rate of sequence variability in the surface (S) gene of HBV because of high rate of mutation due to antiviral therapy.
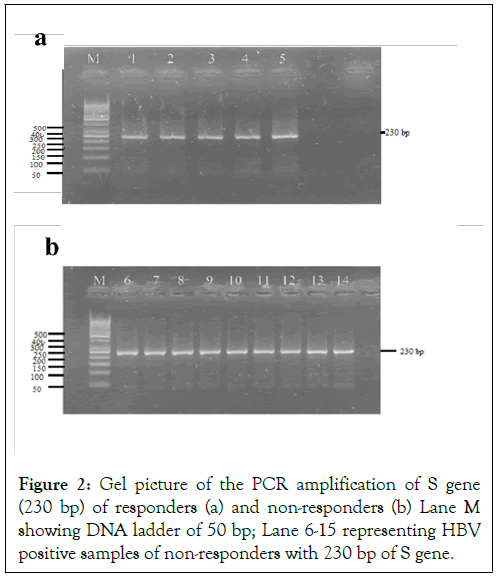
Figure 2: Gel picture of the PCR amplification of S gene (230 bp) of responders (a) and non-responders (b) Lane M showing DNA ladder of 50 bp; Lane 6-15 representing HBV positive samples of non-responders with 230 bp of S gene.
Phylogenetic analysis
Sequencing of HBV S gene was carried out for identification of sequence variability. There were no significant sequence variation among the Responders and non-responders. Nucleotide sequence analysis of the obtained sequences based on HBV S gene sequences with reference sequences from different countries showed that our sequences clustered with some local and regional sequences with high bootstrap values. Bioinformatics tools were used to translate the attained nucleotide sequences to amino acid sequences. To search for alike sequences, the obtained sequences were used using BLAST algorithms against NCBI non-redundant nucleotide and online database of protein. BLAST investigation further established that reported sequences had high resemblance among the responders and non-responders of the surface gene of HBV. Collected sequences were used to build phylogenetic tree by using Maximum likelihood algorithms. Evolutionary analysis revealed that our reported sequences of HBV S gene, clustered with Pakistani isolates. Although they were closely related and evolved together but again were distinct from other countries. The S gene of HBV responders and non-responders was amplified and sequenced. BLAST analysis was performed and S gene sequences having homology with reported sequences were selected randomly for phylogenetic analysis. For finding sequence variation among responder and non-responder, the S gene of 15/150 HBV samples (5 responder, 10 non-responder) was amplified and sequenced. Sequence blasting was also performed between responder and non-responder sequences. Phylogenetic analysis was carried out and a Maximum Likelihood algorithm (1000 bootstrap replicates) tree was developed from the sequences obtained from our study and sequences retrieved from NCBI of various geographical regions of the world (Figure 3). A nucleotide based phylogenetic tree constructed with 30 reported sequences of other countries showed that the sequenced isolates grouped as a separate clade which indicated their relatively close relationship with one another and distinct nature from HBV isolates of the other countries. In nucleotides-based tree by ML method, reported sequences of responders and non-responders were observed. Our sequences clustered together with Pakistani isolates and also showed high homology with the sequences of Saudi Arabia and India with strong bootstrap value. Hence showed that it may be due to trade, travel and communication between these countries. It is suggested that for our sequences dataset, ML tree should be made by using character-based tree to compare our sequences with other countries. In current study the ML tree methods showed and hence verified that our sequences made a distinct cluster in the ML tree and indicated that there is much sequence variability in HBV S gene of Pakistani isolates related to S gene reported sequences from NCBI. Klaus et al. reported that hepatitis B chronic carriers typically indicate HBV surface antigen (HBsAg) in their sera, which is thought the best indicator for mild and chronic infection of HBV. In some individuals, though, routine serological assays cannot detect this antigen in spite of the existence of virus in liver and peripheral blood. The lack of HBsAg might be one of the reasons of mutations in the part of the molecule detected by specific antibodies. The S gene sequences of HBV were determined of samples from 33 virus carriers who were negative for HBsAg but showed antibodies against the virus core (anti-HBc) as the only hepatitis B serological marker. These outcomes recommend that at least some of the chronic low-level HBV carriers, where surface antigen is not identified, might be infected by diagnostic escape mutants and/or by variants with impaired replication. So our study also concluded that as a result of frequent mutation occurs in HBV due to antiviral therapy, there is variation in HBV S gene as our reported sequences made a distinct cluster in phylogenetic tree as compared to S gene sequences of other countries. With reference to this, our study suggested that the surface antigen gene of HBV isolates from KP province showed some similarities with isolates of other countries. No significant variations of nucleotides in the surface gene of HBV was found among the responders and non-responders of antiviral therapy indicating that surface antigen gene may not be important with respect to treatment outcome. Antigenicity of various HBV proteins is determined in order to design more effective vaccines against local strains.
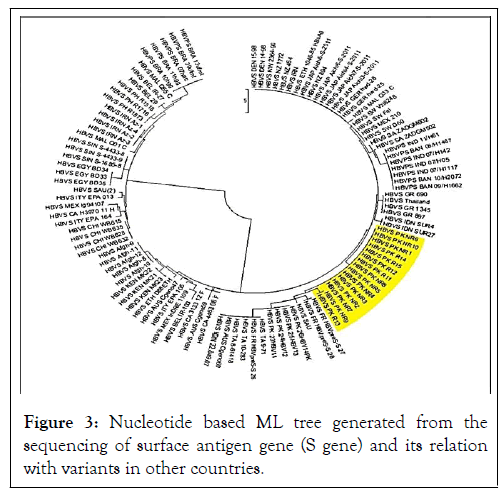
Figure 3: Nucleotide based ML tree generated from the sequencing of surface antigen gene (S gene) and its relation with variants in other countries.
This study reveals that HBV surface antigen gene shows variability in DNA sequence as Pakistani isolates make a distinct cluster in the phylogenetic tree. However, the surface antigen gene of HBV isolates from KP province showed some similarities with isolates of other countries. No significant variations in nucleotides of the surface gene of HBV was found among the responders and non-responders to antiviral therapy indicating that surface antigen gene may not be important with respect to treatment outcome. Hence, the evolutionary analysis of HBV S gene presented important perceptions on its origin, progression and grade of genetic diversity. Phylogenetic analysis on the base of sequences of full genome or more than one gene is necessary for better characterization of this variant and it will be very useful in clinical management.
Consent to publication
Not applicable
Disclosure of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
No funding sources
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Alam N, Daudzai Z, Surareungchai W (2022) Sequence Variability of Hepatitis B Virus (HBV) Surface Antigen Gene (S Gene) among the Non-Responders of Antiviral Therapy. J Hepatol Gastroint Dis. 8:211.
Received: 21-Jun-2022, Manuscript No. JHGD-22-18027; Editor assigned: 24-Jun-2022, Pre QC No. JHGD-22-18027 (PQ); Reviewed: 08-Jul-2022, QC No. JHGD-22-18027; Revised: 15-Jul-2022, Manuscript No. JHGD-22-18027 (R); Published: 22-Jul-2022 , DOI: 10.35248/2475-3181.22.8.211
Copyright: © 2022 Alam N, et al. (2022) is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.