
Cell & Developmental Biology
Open Access
ISSN: 2168-9296

ISSN: 2168-9296
Research - (2020)Volume 9, Issue 4
Coronavirus disease-2019 (COVID-19) has spread globally since its discovery in Hubei province, China in December 2019. It has already been announced as a pandemic disease by world health organization (WHO). This disease is caused by severe acute respiratory syndrome corona virus-2 (SARS-CoV-2), which is an enveloped RNA virus and highly contagious. This is a new coronavirus, still changing, and put the scientific authority in a puzzle. The fatality rate of COVID-19 is higher compared to other coronavirus diseases such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS). The outbreak of COVID-19 has created a severe threat to public health across the world. In this review, we render an overview of COVID-19 disease such as origin and pathogenesis of SARS-CoV-2, clinical manifestations along with complications, diagnosis and treatment actions as well as preventive measures to control viral transmission.
COVID-19; WHO; SARS; MERS; Pathogenesis; SARS-CoV-2
The new coronavirus disease (COVID-19) was first detected in Hubei Province, China in December 2019 [1]. The novel coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first discovered in Wuhan, Hubei Province, China in late December 2019, and subsequently developed a very terrible pandemic. Many patients are admitted to the hospital with fever, cough, shortness of breath and other symptoms. The patient was scanned using computed tomography (CT), which resulted in the initial diagnosis of pneumonia. Further nucleic acid analyses performed using multiple real-time polymerase chain reaction (PCR) of known pathogen groups resulted in negative results, indicating that the cause of pneumonia is unknown [2]. The virus is considered to be a natural virus with animal origin, especially caused by flash food infection [3, 4]. The emergence and rapid spread of this new type of coronavirus SARS-CoV-2 is destroying global health and the economy [5, 6]. To date, SARS-CoV-2 has infected more than 3 million people and caused more than 200,000 deaths. It has forced most regions of the world to adopt a lock-in model, which has caused amazing economic consequences and human suffering [7].
In the past 50 years, various coronaviruses that cause widespread human and veterinary diseases have emerged. Between 2002 and 2003, a new type of coronavirus called SARS broke out in China and Hong Kong, and then spread to other countries such as Vietnam and Canada. Ten years later, a new type of coronavirus called MERS virus (Middle East Respiratory Syndrome) was discovered in Asian countries in the Middle East [8]. It has been discovered that SARS-CoV-2 is related to severe acute respiratory syndrome virus (SARS-CoV), Middle East respiratory syndrome virus (MERSCoV) and bat coronavirus RaTG13, respectively [9,10].
SARS-CoV2 is an enveloped, non-segmented forward RNA virus, contained in the subfamily sarbecovirus, orthocoronavirinae, and widely distributed in humans and other mammals [11, 12]. It has a diameter of about 65-125 nm, contains a single strand of RNA, and has a crowned peak on the outer surface [13, 14]. SARS-CoV-2 can spread from person to person. The current hypothesis is that the first transmission occurred between a bat and an intermediate host animal that has yet to be determined1. It is estimated that a person infected with SARS-CoV-2 will infect about three new people [15]. Common Symptoms include fever, cough, fatigue etc. Symptoms may be similar to those of flu or common cold patients [16]. In case of immune competent individuals, the virus exhibits self-limiting respiratory infections and common colds. But the elderly and immune compromised persons are at risk of getting lower respiratory tract infection and developing pneumonia like symptoms eventually death due to breathing difficulties [17]. The most alarming thing about the virus is its high infectivity and very high rate of transmission.
Currently, there are no therapies or vaccines approved by the US Food and Drug Administration (FDA) for the treatment of COVID-19 patients [18, 19]. However, some pharmaceutical companies, research institutions and universities are trying to discover potential therapies and vaccines against COVID-19 [20, 21]. In this case, the world health organization (WHO) urges the international community to conduct large-scale diagnostic tests to combat the spread of the virus and reduce the number of undetected cases, because the test is also a valuable tool that can help researchers learn epidemiology of the disease. In addition, diagnosis plays a decisive role in deciding the treatment and isolation of infected persons in a timely manner, thereby slowing or preventing the spread of infectious diseases [22]. Although there are currently some diagnostic methods available for virus detection, it is still not possible to diagnose COVID-19 quickly and sensitively. These methods use pathological changes in the patient’s organ by imaging like CT, viral nucleic acid test like RT-PCR, next-generation sequencing of the whole genome, immunological molecules produced by the patient or by the virus in the patient’s body- Antigen–antibody reaction based tests like ELISA [23, 24].
In this review article, we focused on an overview of COVID-19 disease which includes origin, structural and genomic organization of SARS-CoV-2, pathogenesis of SARS-CoV-2, cinical features of COVID-19 patients, diagnostic methods to detect SARS-CoV-2, clinical management and prevention.
Origin of SARS-CoV-2
The origin of the SARS-CoV-2 genome has been linked to bats related to SARS-CoV-1 and MERS-CoV viruses [25]. Interestingly, the whole genome of SARS-CoV-2 is aligned with the viral genomes (Bat-CoV and Bat-CoV RaTG13) of the relative species in Yunnan Province, showing 96% similarity [26]. Some people suspect that inSARS-CoV-2 pangolin is a natural reserve of the virus. This depends on the comparison of SARS-CoV-2 genome contigs, such as SARS-CoV-2 like CoV (Pangolin-CoV) hidden or docked in the lung tissue of two dead Malayan pangolins [27]. The entire genome of the pangolin coronavirus is 91.02% similar to SARS-CoV-2 and 90.55% similar to Bat-CoV RaTG13 (Table 1) [28, 29].
Structure and Genome Organization
| Virus (Disease) | Source of Virus | Transitional Host | Final Host |
| SARS-CoV-1 (SARS-2002) | SARS like Bat-CoV | Civet cat | Human |
| MERS-CoV (MERS 2012) | SARS like Bat-CoV | Camel | Human |
| SARS-CoV-2 (COVID-2019) | Bat-CoV RaTG13 | Pangolin (Pangolin-CoV) | Human |
Table-1: Synopsis of the natural reservoir, median host and target in major coronaviruses.
Structure:
COVID-19 is a single-strand, positive-sense RNA virus enfolded in a lipid bilayer (Figure 1) [30, 31]. The lipid bilayer fuses with the host cell membrane, delivering RNA to the cytoplasm and inoculating the translation of various viral proteins. The replicated RNA genome and artificially made viral proteins coordinate with fresher viruses and burst out of the cell [32, 33]. The SARS-CoV-2 virus is highly sensitive to ultraviolet light and heat [34]. The virus enters through the combination of two proteins. Spike protein (S protein) is a glycoprotein that is expressed in the form of homotrimers on the viral envelope and is called the viral counterpart [35]. Each S protein contains two subunits. The S1 subunit binds to the receptor binding domain that marks the receptor on the host cell, while S2 regulates membrane fusion. This viral S protein binds to the human protein receptor ACE2 [36]. ACE2 is rich in lung, heart, kidney and adipose tissue [37, 38]. The binding of S protein to ACE2 facilitates membrane fusion and the introduction of COVID-19 RNA into cells.
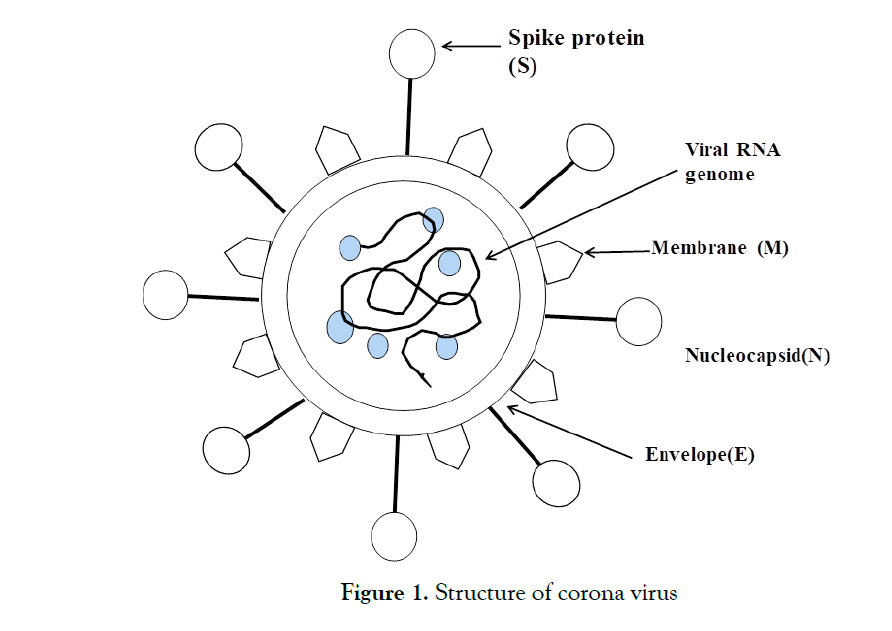
Figure 1: Structure of corona virus.
Genome Organization:
By using codon usage analysis, Wei et al. reported that the virus appeared to be a recombinant virus. However, this view is opposed by Paraskevis's genome-wide evolution analysis and Chen's Simplot survey [39, 40]. According to current research, SARS-CoV-2 is a new type of positive single-stranded RNA virus, belonging to the β-coronavirus genus of the coronaviridae family [41, 42]. This new virus has a genome length from 29,891 to 29,903 nucleotides, which is one of the largest virus among RNA viruses [34]. Similar to SARS-CoV and MERS-CoV, the new SARS-CoV-2 genome has two untranslated regions (UTR), 5'-methylated cap and 3'-poly-A tail structure, and an open reading. The frame (ORF) encodes a polyprotein [43]. At the 5'end, the organization of the SARS-CoV-2 genome is guided by viral replicase (ORF1a and ORF1b). Structural proteins (Spike protein(S), Envelope protein (E), Membrane protein (M) and Nucleocapsid (N)) and at the 3′ end some genes of accessory proteins, such as ORF 3a, 7, and 8, are arranged in genes of structural proteins (Figure 2) [44-46].
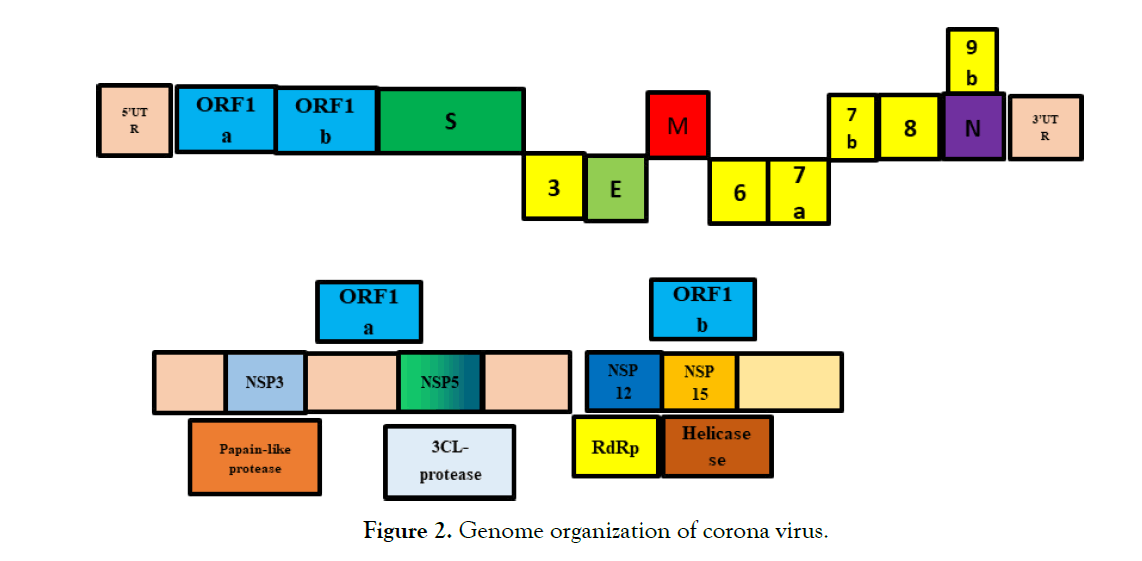
Figure 2:Genome organization of corona virus.
In the genome of the coronavirus, the genes of ORF1a and ORF1b account for about two-thirds of the entire genome, encoding 16 non-structural proteins (nsps), while the remaining one-third encode accessory proteins and structural proteins . Eight complete and two partial or incomplete genome sequences for SARS-CoV-2 were obtained by NGS method.
Life cycle
Here are some of the key steps involved in the infection of host cells by coronavirus (Figure 3).
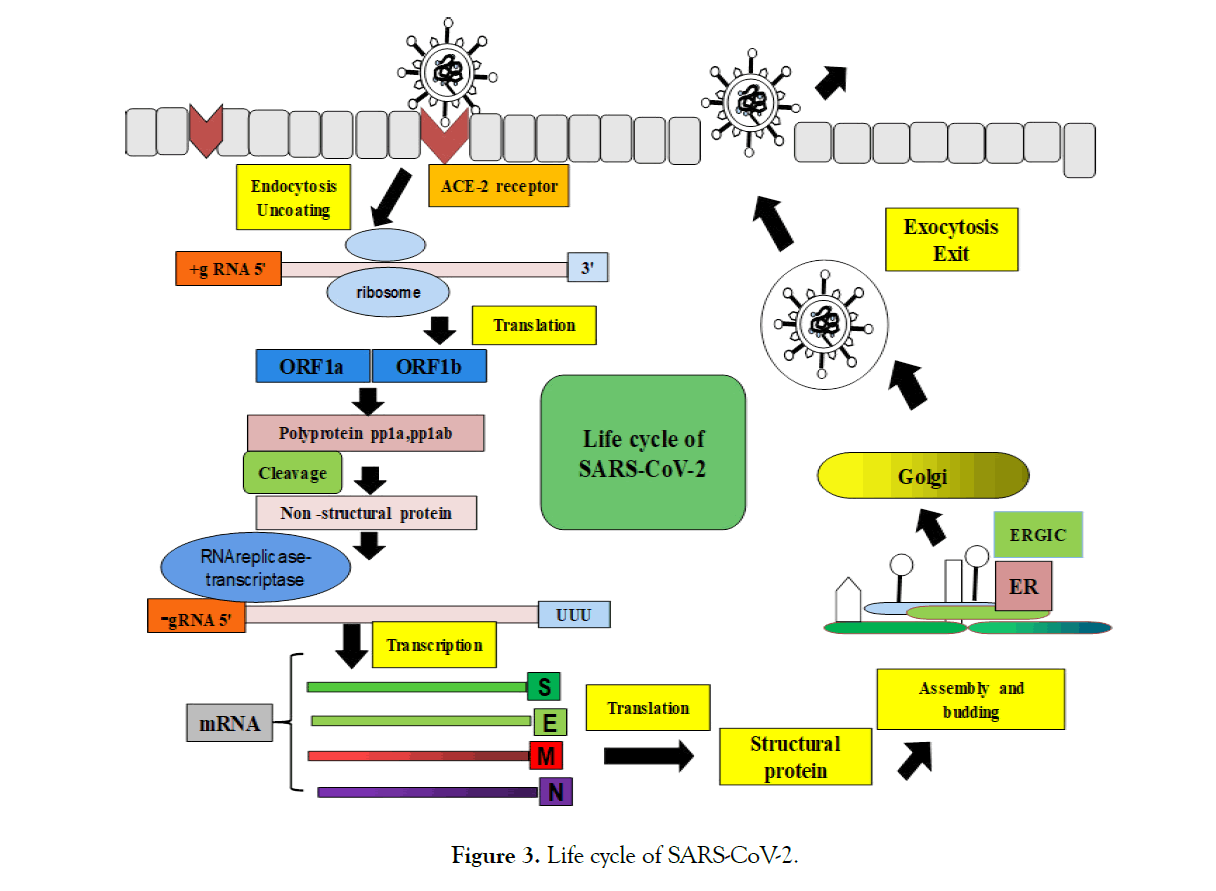
Figure 3:Life cycle of SARS-CoV-2.
1. The virus attaches to the target host cell and enters the cell through endocytosis
2. Use host cell translation mechanism to translate and express replicase protein from viral genome RNA
3. RNA replication and transcription into mRNA
4. The viral protein assembly after translation and release by exocytosis
Compared with SARS, COVID-19 uses the same mechanism to enter host cells, but at a slower rate. COVID-19 is still accumulating more in the system compared with SARS. This explains why the incubation period of COVID-19 is longer and more infectious, while SARS appears with more symptoms and a tighter disease [47, 48]. SARS-CoV-2 may be able to enter through novel clathrin- and caveolae-independent endocytic pathways together with clathrindependent entry mechanisms, although the exact mechanism by which SARS-CoV-2 enters into the cells may depend on the cell type. Of cells, different results have been found [49, 50]. SARSCoV- 2 may enter the cell through the endocytic pathway or directly fused with the plasma membrane. SARS-CoV-2 can bind to the ACE2 receptor, as well as the sialic acid residues on gangliosides [51].
Transmission of SARS-CoV-2
The first case was direct contact with infected animals (transmission from animals to humans) at the seafood market in Wuhan, China. However, there have been clinical cases with different exposure history. Person-to-person communication is now regarded as the main form of communication. Individuals who seem to be asymptomatic may also carry the virus. However, the most obvious source of transmission is people with symptoms. Transmission occurs through the spread of respiratory droplets by coughing or sneezing. The data also shows that close interaction between individuals may also lead to transmission. Due to the increased aerosol concentration, this also suggests that it may spread in enclosed spaces. The basic copy number of SARS-CoV-2 is 2.2, and in some cases 3 or more [24, 34]. This informs the patient that the infection can be spread to two other people. Available data indicates that the virus has an incubation period of three to seven days, but some data has shown that the virus has an average incubation period of 5 days and an incubation period of 2-14 days [52, 53].
These results are based on the initial case. Therefore, further research is needed to solve the propagation kinetics and incubation time [54]. The mode of transmission found in imported cases are through droplet transmission, fecal to oral route, conjunctiva and poisonous gas [55, 56]. In addition, local transmission can be traced back to the patient's body fluids, such as respiratory droplets, saliva, feces and urine. Virions are fixed at a lower temperature that is 4°C, has a higher survival rate than 22°C [57, 58]. As SARS-CoV- 2virions are cast throughout the clinical course, patients infected with COVID-19 can spread the infection before the symptoms appear, during the symptoms, and during the clinical recovery. Additional consideration must be given to the residence time of SARS-CoV-2 virions on the surface. According to some data obtained through continuous research, the half-lives of SARSCoV- 2 virus on copper metal, aerosol, cardboard, stainless steel and plastic surfaces are 1, 1.5, 3.4, 5.6 and 6.8 hours, respectively. The effective retention period of SARS-CoV-1 in aerosol, copper, cardboard, stainless steel and plastic is 3h, 4h, 24h, 48h and 72h respectively [59].
Host response
Although, receptor recognition is not the only determinant of species specificity. After binding to the receptor, SARS-CoV-2 will immediately be inserted into the host cell where it encounters an innate immune response. In order to effectively infect new hosts, SARS-CoV-2 must be able to suppress or evade the host's innate immune signals.However, how SARS-CoV-2 evades immune response and causes pathogenesis is almost unknown. Considering that COVID-19 and SARS have close clinical features [60]. The pathogenesis of SARS-CoV-2 may be similar to SARSCoV. Regarding SARS-CoV infection, the type I interferon (IFN) system can promote the expression of IFN-stimulating genes (ISG), thereby inhibiting virus replication. In order to overwhelm this antiviral activity, SARS-CoV encodes at least 8 viral antagonists, which balance the induction of IFN and cytokines and the function of hedging ISG effectors [61]. By mediating inflammation and cellular antiviral activity, the host immune system's response to viral infection is critical to hinder virus replication and spread. However, excessive immune response and virus lysis on host cells will result in pathogenesis. Studies have shown that patients with severe pneumonia usually present with fever and dry cough at the onset of sickness.Some patients develop rapidly due to acute respiratory stress syndrome (ARDS) and septic shock, eventually leading to multiple organ failure, and about 10% of patients die [62]. The development of ARDS and severe lung damage in COVID-19 further indicate that ACE2 may be the entry route of SARS-CoV-2, because ACE2 is abundant in human airway epithelial ciliated cells and type II alveolar lung cells [63]. Patients with SARS and COVID-19 have related forms of inflammatory damage. In serum from patients diagnosed with SARS, there is elevated levels of pro-inflammatory cytokines (e.g. interleukin (IL- 1, IL-6, IL-12, interferon gamma, IFN-γ-induced protein 10 (IP10), macrophage inflammatory proteins1A (MIP1A) and monocyte chemo attractant protein-1 (MCP1) which are associated with lung inflammation and severe lung injury [64]. In addition, patients infected with SARS-CoV-2 are described to have higher plasma levels of pro-inflammatory cytokines such as IL1β, IL-2, IL7, TNF-α, GSCF, MCP1 than normal adults. Eminently, patients in the intensive care unit (ICU) have a remarkably higher level of GSCF, IP10, MCP1, and TNF-α than those non-ICU patients, indicating that a cytokine storm might be an underlying root of disease severity.Unusually, anti-inflammatory cytokines such as IL10 and IL4 were also raised in those patients [60], which was unusual phenomenon for an acute phase viral infection. Study suggests that, an elevated level of three distinct cytokines are surprisingly negatively correlated with the whole of T cell counts, CD4+ counts, and CD8+ counts, respectively. Hence lower number of T cells can be an indication to a new therapeutic measures in early disease progression [65]. Another interesting finding was that SARS-CoV-2 has shown to specially infect older adult males with rare cases noted in children [62]. The same direction was observed in primate models of SARS-CoV where the virus was observed more likely to infect aged Cynomolgus macaque than young adults [66]. Additional studies are necessary to determine the virulence factors and the host genes of SARS-CoV-2 that allows the virus to transverse the species-specific barrier and cause deadly disease in humans [67].
The clinical manifestations of COVID-19 infection are similar to SARS-CoV, and the most common symptoms include fever, dry cough, dyspnea, chest pain, fatigue and myalgia. Less common symptoms include headache, dizziness, abdominal pain, diarrhea, nausea and vomiting [68-70]. According to the report of the first 425 confirmed cases in Wuhan, common symptoms include fever, dry cough, myalgia and fatigue, and less common symptoms are sputum production, headache, hemoptysis, abdominal pain and diarrhea [71]. Approximately 75% of patients have bilateral pneumonia [72]. However, unlike SARS-CoV and MERS-CoV virus infections, few patients with COVID-19 have significant upper respiratory tract signs and symptoms, such as nosebleeds, sneezing or sore throat, which indicates that the virus may be more prone Infects the lower respiratory tract of the lung [68]. Pregnant women and nonpregnant women have similar characteristics. Severe complications and complications have been noted in COVID-19 patients, such as hypoxemia, acute ARDS, arrhythmia, shock, acute heart injury and acute kidney injury. A study conducted in 99 patients found that approximately 17% of patients developed ARDS, and 11% of patients died of multiple organ failure [73]. The median time from initial symptoms to ARDS is 8 days. The mortality rate of COVID-19 ranges from 0 to 14.6% [74]. Although, Yang et al. According to reports, of 52 severely ill and intensively ill adult ICU patients, 32 (61.5%) died at 28 days [75]. It is not difficult to see that the severity of the disease is an independent predictor of poor prognosis [76]. The analysis of non-ICU patients showed that ICU patients were older, had more comorbidities, dyspnea, abdominal pain, and anorexia symptoms were more frequent. At the same time, there are reports that ICU patients have higher levels of plasma cytokines and chemokines, IL2, IL7, IL10, GSCF, IP10, MCP1, MIP1A and TNFα [77, 78]. Non-survivors have more acute lymphopenia and higher blood cell counts, neutrophil counts, d-dimers and fibrin impairment products than survivors [79]. Overall, the mortality rate of SARS-CoV-2 is slightly lower than that of SARS-CoV and MERS-CoV (9.6% and 40.0%, respectively) [80, 81].
Testing has become a key part of the response to the COVID-19 pandemic. In addition to the gold standard PCR test used to detect currently infected patients, many auxiliary testing procedures are also under development. Scientists have been working in the universe to increase the availability of coronavirus testing to combat the COVID-19 pandemic.
Diagnosis plays a vital role in the containment of COVID-19, so that control measures can be quickly implemented to expand the scope of transmission through case identification, isolation and contact tracing. COVID-19 is available with two detection methods: virus detection and antibody detection test. A viral test tells us if a patient has a current infection and the antibody test tells us if we have ever been infected. Antibody tests may not show whether we are infected with the virus, because it may take 1-3 weeks for the body to produce antibodies.
If an individual tests positive or negative for COVID-19 in a virus or antibody test, he/she should still take precautions to protect himself and others. Diagnostic tests can show whether a suspected case has an active coronavirus infection and measures should be taken to isolate or isolate it from other people. Diagnostic tests are also called virus tests, molecular tests, nucleic acid amplification tests (NAAT), RT-PCR tests, and LAMP tests.
Nucleic acid testing
Nucleic acid test is the first-hand method to diagnose COVID-19 in the diagnosis of genetic material of SAV-CoV-2 [82].
RT-PCR
RT-PCR is the most important method for identifying COVID-19 using respiratory samples. Many reverse transcription polymerase chain reaction (RT-PCR) kits have been configured to detect SARS-CoV-2 genetically. RT-PCR refers to reverse transcription of SARS-CoV-2 RNA into complementary DNA (cDNA) strands, and then amplification of specific regions of cDNA [83-86]. System operation usually includes two main steps: (1) sequence alignment and primer design, and (2) analysis optimization and testing.
Genome sequencing is essential for researchers to design primer and probe sequences for PCR and different nucleic acid tests compared and analyzed the number of viral genome sequences related to SARS and designed a set of primers and probes [87]. They observed three regions with conserved sequences: A. RdRP gene (RNA-dependent RNA polymerase gene) in the ORF1ab region of the open reading frame, B. the E gene (envelope protein gene) and C. the N gene (nucleocapsid protein) gene).Both RdRP and E genes have wide detection sensitivity, while N gene shows poor analytical sensitivity. The detection can be configured as two target systems where one primer can usually detect many coronaviruses, including SARS-CoV-2, while the second primer can only detect SARS-CoV-2. The primers and probes are then designed, and the next steps involve optimizing assay conditions, such as reagent conditions, incubation time and temperature, and then performing PCR testing. RT-PCR can be performed in one-step or two-step tests. Finally, the control needs to be carefully selected to ensure the reliability of the measurement and to determine experimental errors. RT-PCR is a diagnostic assay using nasal swabs, tracheal aspirates, or bronchoalveolar lavage specimens. The main method of diagnosis is the accumulation of upper respiratory tract samples through nasopharyngeal and oropharyngeal swabs. It is not recommended to use bronchoscopy as a diagnostic method for COVID-19, because the generated aerosol poses a considerable risk to both patients and medical staff. In a case study series, Zou et al. observed that SARS-CoV-2 RNA levels were high in samples collected from the upper respiratory tract and the first 3 days after the onset of symptoms, and SARS-CoV-2 RNA was also found in samples collected from the upper respiratory tract of asymptomatic patients [88]. Various reports indicated that SARS-CoV-2 RNA can also be detected in blood and stool samples [89-92]. The specificity of the RT-PCR test appears to be very high, whereas there may be false-positive results due to swab contamination, particularly in asymptomatic patients. The sensitivity is not clear, but it is about 66–80%. RT-PCR depends on whether there is detectable SARSCoV- 2 in the collected samples. If an asymptomatic patient is infected with SARS-CoV-2 but is cured, PCR will not be able to determine the previous infection, and control measures will not be implemented [93].
Isothermal amplification
The nucleic acid test using isothermal amplification is currently being used for SARS-CoV-2 detection. The isothermal amplification method is performed at a single temperature and does not require technical laboratory equipment to provide similar analytical sensitivity to PCR. These methods include recombinase polymerase amplification, helicase-dependent amplification and loop-mediated isothermal amplification (LAMP). Different academic research laboratories have formulated and performed clinical reverse transcription LAMP tests for SARS-CoV-2 [94- 98]. RT-LAMP uses DNA polymerase and 4 to 6 primers to bind to 6 defined positions on the target genome. In the four-primer system, there are two inward primers (forward and reverse internal primers) and two outward primers. LAMP is very unique because it uses a large number of primers. In the LAMP diagnostic run, the patient sample is placed on a test tube and the amplified DNA is identified by turbidity (a byproduct of the reaction), color (inclusion of a pH-sensitive dye), or fluorescence (addition of a fluorescent dye that binds to double-stranded DNA). The reaction is carried out at a temperature of 60-65°C for less than 1 hour, and the detection limit of the analysis is about 75 copies per μL. The test procedure is simple to operate, easy to observe for detection, has fewer background signals, and does not require a thermal cycler. The disadvantage of LAMP is the challenge of optimizing primers and reaction conditions. Other isothermal amplification techniques for COVID-19 detection are under development [99, 100]. Isothermal amplification techniques can be diverse in the amplification and/or readout stages. Multiplexing can use polymer beads encoded with specific optical markers (such as organic fluorescent molecules) for barcodes. It is possible to organize barcodes for different biomarkers in the panel to detect multiple analytes from a single patient sample in a reaction tube [101].
CRISPR technology
To facilitate the diagnosis of COVID-19, hot topics research has implanted a protocol for diagnosis using CRISPR-based technology. The CRISPR-based diagnostic kit is made up of Sherlock Biosciences, a biotechnology company based in Cambridge. It works by programming the CRISPR mechanism, which has a certain gene sequence resident ability and can detect SARS-CoV-2 genetic material in nose, mouth or throat swabs or lung fluid. If the genetic material of the virus is found, the CRISPR enzyme will fluoresce. According to the organization, the test can restore results in about 60 minutes. The CRISPR platform uses nucleic acid biomarkers and RPA technology for CRISPR/Ca9-mediated lateral flow nucleic acid determination (CASLFA) and PCR of serum samples [102]. Another platform uses nasopharyngeal swab samples for RT-RPA method, showing RPA, SHERLOCK detects multiple signals by fluorescence [103]. Quantum dot platform barcodes use nucleic acid labels and serum samples, in which multiple quantum beads capture viral DNA for RPA detection [63]. Magnetic beads, paramagnetic beads, and magnetic bead isolation are some examples of magnet-based detection systems [104-106].
Computed Tomography
Chest CT scan is non-invasive and requires multiple X-ray measurements at various viewpoints of the patient's chest to create a cross-sectional image. Radiologists analyze these images to look for atypical features that may lead to diagnosis [107, 108]. The imaging features of COVID-19 are diverse and depend on the stage of infection after the onset of symptoms. The most popular marker features of COVID-19 include bilateral and peripheral ground-glass opacity and pulmonary consolidation found that the turbidity of the glass wool was most obvious 0 to 4 days after the onset of symptoms. With the development of COVID-19 infection, in addition to the opaque glass, it will also form a crazy form of paving, and then increase the lung consolidation [109-111]. Based on these tomographic characteristics, various retrospective studies have shown that, compared with RT-PCR, CT scans have higher sensitivity (86-98%) and a higher false negative rate [112-115]. The main warning of using CT for COVID-19 is low specificity (25%), because imaging features converge with other viral pneumonias; on the other hand, CT systems are expensive, require specialized technical knowledge and cannot specifically diagnose COVID-19.
Protein Testing
The viral protein antigens and antibodies produced by SARSCoV- 2 infection can be used to diagnose COVID-19. Changes in viral load during the course of infection may result in viral proteins that are difficult to detect. For example, It was found that the saliva viral load is high in the early stage after the onset of symptoms and gradually decreases over time . In order to modify the surveillance attempt, serological testing of protein is also required as well as nucleic acid test. Unlike nucleic acid tests, these assays have the benefit of sensing after recovery. This allows doctors to track sick patients and recovered patients, so as to better calculate the total infection rate of SARS-CoV-2 [116, 117]. Currently, serological tests for specific antibodies, such as blood tests are under development identified immunoglobulins G and M (IgG and IgM) in the serum of human coronavirus-positive patients by enzymelinked immunosorbent assay (ELISA). They used the SARS-CoV-2 Rp3 nucleocapsid protein, whose amino acid content is 90% similar to other SARS-related viruses. If anti-SARS-CoV-2 IgG is present, it will be sandwiched between the adsorbed nucleoprotein and anti-human IgG probe and generate a positive signal. The IgM test done by Zhang et al. has a related structure but uses antihuman IgM adsorbed to the plate and an anti-Rp3 nucleocapsid probe. They tested 16Covid-19 positive patient samples (affirmed by RTPCR) and found the levels of these antibodies enhanced over the initial 5 days after symptom onset . Antibodies were found in the respiratory tract, blood or stool samples. Xiang et al. recognized SARS-CoV-2 IgG and IgM antibodies in suspicious cases [118-120]. Guan etc. showed that the levels of C-reactive protein and D-dimer in infected patients increased and the levels of lymphocytes, white blood cells and platelets decreased [113]. The challenge in using these biomarkers is that they are also abnormal in other diseases. Multiple tests using antibodies and small molecule markers at the same time can change the specificity. ELISA, SIMOA and biobarcoding assays use protein biomarkers. ELISA performs an enzymatic reaction in the presence of the target serum to produce a colored product. SIMOA uses a digital version of ELISA for target samples. Smartphone dongle platform uses a microfluidic-based cassette. ELISA for blood sample bio-barcode determination using DNA-assisted immunoassay indirect detection. Protein signals are detected by amplifying DNA bound to gold nanoparticles [121- 125].
Point-of-Care Testing
Point-of-care testing is a cost-effective handheld device used to diagnose patients outside of centralized facilities. These can be operated in areas such as community centers to reduce the burden on clinical laboratories [116]. The lateral flow antigen test for SARS-CoV-2 is an improving point of care method for diagnosing COVID-19 [119]. Rapid antigen detection is a protein-based pointof- care detection system that uses a lateral flow method in which gold-coated antibodies create a colorimetric signal on paper in the presence of the target [121]. Lateral blood flow analysis showed that the clinical sensitivity, specificity and accuracy of IgM were 57%, 100% and 69% for IgM, and 81%, 100% and 86% for IgG, respectively. The clinical sensitivity of a test that can detect both IgM and IgG is 82% [119]. Another application used in the medical field is microfluidic devices. These devices consist of micron-sized channels and palm-sized chips in the reaction chamber. The chip uses electric, capillary, vacuum and other forces to combine and separate liquid samples. These chips can be made of materials such as polydimethyl sulfoxide, glass or paper. The main advantages of using microfluidic technology include miniaturization, small sample size, fast detection time and portability [126]. These applications can be modified to detect SARS-CoV-2 RNA or protein. The focus is on many emerging technologies that can be used to detect SARS-CoV-2. Academic research laboratories are developing many platforms, such as electrochemical sensors, paper-based systems and surface-enhanced Raman scattering-based systems. These methods are still in the early stages of development and cannot be used to diagnose COVID-19 immediately. These emerging technologies may play a role in detecting an impending disease.
Role of smartphones in diagnostics
Smartphone features (such as connectivity, database infrastructure and onboard hardware) can improve evidence-based policy development, national disease response coordination, and community healthcare. Controlling of epidemics and pandemics requires extensive surveillance, epidemiological data sharing and patient monitoring [127,128]. Health care entities from general hospitals to the WHO need tools that can improve the speed and convenience of communication to manage the spread of disease. Smartphones can be invested for this purpose because of their connectivity, computing power, and hardware that can simplify electronic reports, epidemiological databases, and instant inspections [129-131].
Clinical management and treatment
Currently, there are no certified drugs for the treatment of the COVID-19 pandemic, and vaccines cannot be purchased [132- 134]. Clinical management is mainly based on supportive therapy and treatment of symptoms of viral diseases, and ultimately prevention of respiratory failure. Self-isolation at home is the best choice for patients with mild cases. These patients should maintain adequate water and nutrition and treat indications such as fever, sore throat or cough. Therefore, patients with severe illness can use hospital beds [135, 136]. In view of antiviral, anti-inflammatory and immunomodulatory drugs, cell-based therapies, antioxidants and different clinical preliminary studies of smart drugs against the new coronavirus are being studied [137].
Antiviral Therapy
Attempts have been made to use antiviral drugs such as antiviral drugs, remixivir, lopinavir-ritonavir, depend on anecdotal data on HIV, respiratory diseases, and MERS infection therapy [138,139]. However, the use of antiviral drugs should be prevented under conditions of comorbidities and high mortality risk. As a nucleotide analog, Remdesivir works by being incorporated into the ribonucleic acid strand of the born microorganism, which then causes its premature termination. It has been reported that Remdesivir has been transferred in preclinical studies of SARSCoV and MERS-CoV infection by acting on the infectious agent of coronavirus [140]. North american study of MERS-CoV in mice has shown that Remdesivir is effective in reducing the load of infectious agents and improving the functional parameters of the respiratory organs [141]. The effectiveness of lopinavir/ritonavir against SARSCoV is undisputed [142], and these medications additionally appear to cut back the infective agent load in COVID-19 patients [143,144]. Antibiotics and/or antifungals are needed if co-infections, such as Mycoplasma and Chlamydia, are suspected or proven. Extended macrolide therapy, as a modulator of immune function, is being evaluated [145].
Anti -malarial and anti-parasitic drugs
Chloroquine and hydroxychloroquine are used for the remedy of protozoal contamination and amebiosis. various studies have incontestible antimalarial drug interest in vitro and in animal models towards SARS-CoV [146,147] and craniate grippe [148] . Yao et al. confirmed that, in vitro, anti inflammatory drug is more potent than antimalarial drug in inhibiting SARS-CoV-2 but it is a few not unusual place side results and arrhythmogenic cardiotoxicity [149,150]. Finally, it seems that antimalarial drug medications may want to act synergistically with macrolides (e.g. azithromycin) for increased antiviral result [151].
It has been shown that the antiparasitic drug ivermectin can inhibit the replication of SARS-CoV-2 in vitro. It was previously found that, 2 hours after SARS-CoV-2 infection, Ivermectin in VerohSLAM cells has extensive antiviral activity in vitro and can be used as an inhibitor of pathogen viruses. It is suitable for reducing viral ribonucleic acid replication by about 5000 times within 48 hours. Ivermectin seems necessary to further study the achievable benefits of COVID-19 disease [152].
Oxygen support and ventilation
If there is hypoxia (SatO2 <93%) or symptoms of respiratory distress, oxygen therapy is required. Once the vascular oxygen level has not been reached (SatO2 93-96%), and if acute respiratory damage occurs, invasive mechanical ventilation and intubation are required [153]. Extracorporeal membrane oxygenation may be a feasible treatment for COVID-19 patients who are plagued by severe respiratory diseases [154]. When steroid-based medical treatment becomes necessary, it is necessary to use rock bottom feasible doses, and only for a short time, because the use of steroids to treat SARS-CoV and MERS-CoV cases is associated with many complications [155-158].
Anti -inflammatory and immunomodulatory molecules
People are considering the use of a variety of monoclonal antibodies and immunostimulants to neutralize variants such as SARS-CoV-2 [159]. The use of specific anti-inflammatory molecules such as tocilizumab (anti-IL-6R antibody) is of increasing interest. Anti- IL-17, antiviral agents (such as interferon) and mesenchymal stromal cells for the treatment of severe respiratory diseases (called "acute respiratory distress syndrome") are different potential drug therapies [160]. Expansion of anti-covid19-specific T lymphocytes may also be another possible option [161].
ACE Inhibitor (ACEi) and different blockers
Inhibitors of the connection between the spike (S) protein of the virus and ACE2 [162-164], angiotensin receptor one blockers (sartanics) [165], emodin, promazine [166,167], furin (an amino acid endoprotease) and monoclonal antibodies against the S1 domain of the S protein [168]. An exciting strategy would be targeting the structural genes for the S protein or envelope or membrane proteins with tiny meddling RNAs.
Steroids
Steroids are associated with a higher risk of expiration in influenza patients and delayed virus clearance in patients infected with MERSCoV. Although glucocorticoids are commonly used to treat SARS, there is no good description of their efficacy, but there are convincing data on short-term and long-term harmful effects. The effects of steroids have not been proven and may have a slanting effect, and there is a positive consensus around the world recommending not to use them. The WHO/CDC recommends not to use these drugs in patients with COVID-19 complicated with pneumonia except for other signs (for example, exacerbation of chronic obstructive pulmonary disease and asthma) [169]. The Chinese guidelines also advocate the use of low- and medium-dose steroids for the treatment of ARDS complications of COVID-19 disease [170]. The use of corticosteroids should depend on the intensity of the inflammatory response, the degree of dyspnea (with or without ARDS or without ARDS), and the progress of lung imaging. Corticosteroids can be used in a short time. The recommended dose of methylprednisolone should not exceed 1-2 mg/kg/day. Dexamethasone is a cheap and commonly used steroid. It can be proved by a randomized controlled clinical trial in the United Kingdom to save the lives of severely ill patients with COVID-19. In this trial, it reduced the deaths of patients using ventilators due to coronavirus infection by about a third [171].
Convalescent plasma and antibody therapy
Convalescent plasma is one of the achievable treatment methods for COVID-19. Convalescent plasma is plasma derived from cured patients with COVID-19 which contains SARS-CoV-2 antibodies and can be transfused to infected patients to speed up the disease. Rehabilitation plasma therapy has been used to treat various diseases from measles to polio, chickenpox and SARS for more than 100 years. When necessary, intravenous immunoglobulin can be used for severe COVID-19 disease, but its efficacy is still questionable and further research is needed [172,173]. If IgG antibodies are collected from cases with improved SARSCoV-2 infection to increase the chance of inactivating the virus, the effect of IVIG may be better. By enhancing the immune response of infected cases, more specific IgG antibodies will be more effective against COVID-19 disease [174,175]. Therefore, immunotherapy with special IgG antibodies and antiviral drugs can become an alternative therapy for COVID-19 disease until a suitable vaccine (such as a vaccine) can be selected. It is the first report that the SARSCoV specific human monoclonal antibody CR3022 can effectively bind to the SARS-CoV-2 receptor binding domain. Researchers have published reports of specific monoclonal antibodies against to COVID-19 (B38, H4, 47D11) and hope that this method is effective. The US National Institutes of Health (NIH) has initiated a Phase III clinical trial, named ACTIV-3, to evaluate various types of monoclonal antibodies as potential treatments for hospitalised Covid-19 patients. Strong binding of SARSCoV-2 spike protein by a SARSCoV specific human monoclonal antibody could be an optional therapeutic approach in the near future [176].
Interferon therapy and cytokine storm inhibitor
Interferon (IFN)-α can reduce the viral load in the initial stage of COVID-19 disease, and can help improve disease performance and limit the infection process. The IFN-α atomized drug is dissolved in sterile water and used twice a day for 5-7 days. IFN-α 2b inhalation therapy can be performed on high-risk individuals who are in close contact with presumed SARS-CoV-2 infection cases or individuals with only the early stage of upper respiratory tract manifestations [177]. Excessive inflammation is considered to be one of the most critical negative prognostic indicators in COVID-19 disease. It is caused by a cytokine storm caused by an enhanced immune response to SARS-CoV-2. If a cytokine storm occurs, it is considered helpful to use an exclusive cytokine blocker (such as anakinra or tocilizumab). Anakinra is one of the interleukin antagonists, which can block the results of interleukin-1, in the cytokine storm [178]. The plan of the National Health Commission of China includes the IL-6 inhibitor tocilizumab for the terrible COVID-19 cases and high IL-6 levels during the cytokine storm phase of the COVID-19 disease, which is being evaluated in clinical trials [179].
Prevention
The COVID-19 is the third novel coronavirus to cause a broadscale epidemic in the twenty-first century after the attack of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) in 2003 and theMiddle East Respiratory Syndrome Coronavirus (MERSCoV) in 2012 [180-184].
Preventive measures must focus on optimizing infection control protocols, self-isolation, and patient isolation during the provision of clinical care. The WHO has advised against close contact with patients, farm animals, and wild animals [180]. Patients and the public must cover coughs and sneezes to prevent the spread of aerosols. One should need to wash his/her hands often with soap and water. As an alternative, hand sanitizer can also be used. Individuals with low immunity are advised to avoid public gatherings. The emergency department must adopt strict hygiene measures to control infection. Medical staff must use personal protective equipment, such as N95 masks, FFP3 masks, work clothes, goggles, gloves and work clothes.
Self-Protection
One should wash his/her hands for at least 20 seconds before going to an open place. It is recommended to use soap or hand sanitizer with at least 60% ethanol. It is also recommended to avoid touching the facial area (eyes, nose, mouth), as this is the entry point for the virions into the upper respiratory tract. Everyone’s should avoid interacting with people who are already showing symptoms, and also should avoid gathering in crowded places. Travel to the outbreak area must be prohibited. Healthy people must keep at least three to six feet away from people with symptoms. It is beneficial to disinfect frequently treated surfaces. All medical staffs managing COVID-19 patients need a full set of personal protective equipment (PPE), which includes surgical masks, double gloves, full-sleeve surgical gowns and eye sheild. Before performing procedures which have higher risks related to aerosol exposure such as tracheal intubation, bronchoscopy, cardiopulmonary resuscitation and non-invasive ventilation, the N95 mask must be thoroughly broken to prevent 95% of the droplets from entering the mask. These steps may nebulize the virus. Community transmission can be suppressed by closing educational institutions, businesses, airspace and sports events. High-risk groups (such as those over 65 years of age or those with chronic comorbidities but without any symptoms) must also selfisolate to reduce the possibility of COVID-19 contraction [181].
Herd Protection
After the onset of any symptoms, potential patients should selfisolate and be isolated in a separate room with a separate bathroom for at least 14 days. This self-isolation must also be extended to pets because of documented cases of human-to-dog transmission. If there are other concerns about COVID-19, it is necessary to establish a public health hotline via telemedicine or a general clinic for immediate contact for potential diagnosis. Patients with COVID-19 need to wear a mask (N95) to prevent the spread of droplets [182].
In most cases, public health measures are essential to manage the spread of COVID-19. If the public health measures for containment are insufficient, the patient burden will exceed the available ICU beds and mechanical ventilation capacity. Therefore, the entire goal of COVID-19 management is to suppress the rapid emergence of new cases in a short period of time under the premise of social isolation. This epidemiological concept is called "curve flattening". The ministry of public health should identify infected cases, isolate these cases, conduct contact tracing and isolate symptomatic contacts.
The spread of COVID-19 be minimized or prevented by following guidelines provided through WHO for mass people (Figure 4).
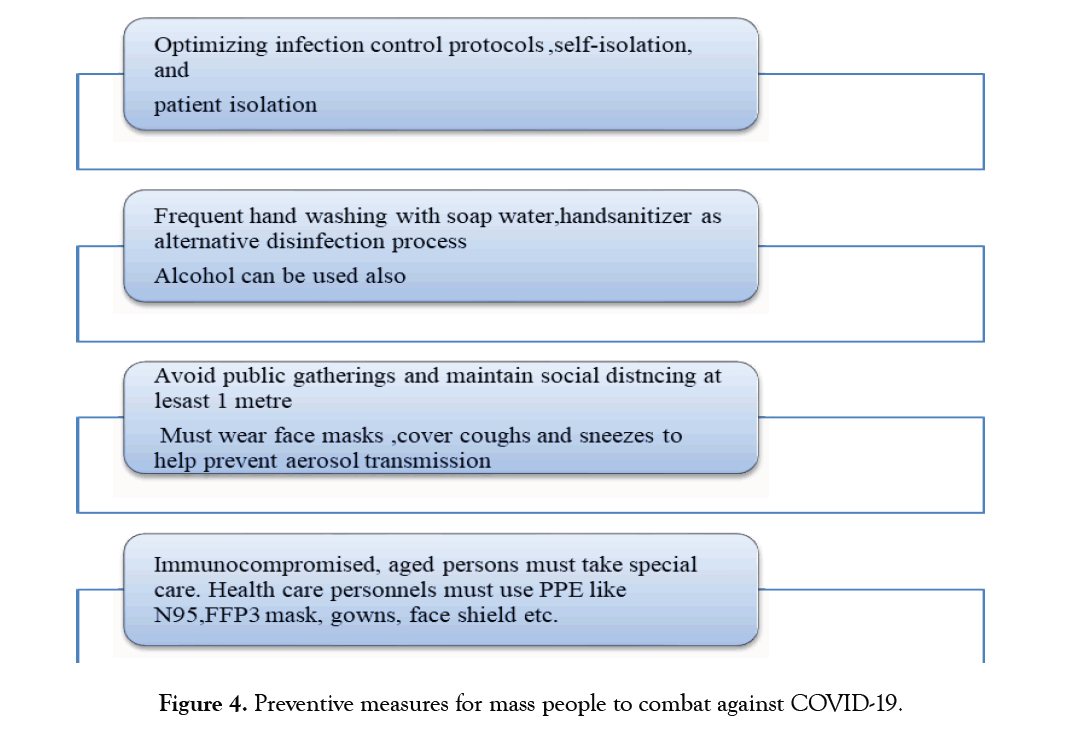
Figure 4:Preventive measures for mass people to combat against COVID-19.
Boosting immunity
A balanced diet, oral health, proper exercise, regular rest, avoiding excessive fatigue and enhancing immunity are powerful measures to prevent various infections (such as virus attacks). It is also important to maintain emotional stability and mental health. Regular intake of vitamin supplements (such as vitamin C and D) is very effective against viral infections [183]. Vaccination is an effective way to prevent viral infections. At present, research and development of antiviral vaccines have been carried out in different countries.
The current COVID-19 pandemic is clearly a general crisis. The rapid increase in the number of infected cases and deaths puts the international community at risk. SARS-CoV-2 seems to be more contagious than SARS-CoV or MERS-CoV and causes more deaths. Most infected individuals with no or mild symptoms can release the virus and spread the virus to others, which is extremely challenging to stop the spread of COVID-19. The only interventions that are currently feasible and proven to be effective in this crisis seem to be maintaining strict social distancing measures and personal hygiene. In addition, nutritional supplements, symptomatic treatment and antiviral treatment are essential for both mild and severe patients. There is no effective vaccine against COVID-19. Proper vaccination is required to prevent emerging coronavirus-related epidemics or pandemics in the future. Now, the best action is to develop a vaccine to prevent infection. Some potential candidate vaccines have entered phase I and II clinical trials, but it may take a year and a half before effective vaccines can be reviewed through trials and ready to be put on the market. Therefore, great efforts should be made to limit the spread of the virus. In addition, the pandemic will simultaneously generate demand for medicines and vaccines on a global scale. The elderly and people with underlying diseases or chronic comorbidities are at greater risk of serious illness or death. Once an effective treatment or vaccine is obtained, clinical and serological studies are needed to confirm which populations are still at the highest risk. Research, pharmaceutical companies, regulatory agencies and governments require strong international coordination and cooperation to ensure the successful manufacture and supply of promising therapies or vaccines. It is time for us to work together, exchange experiences, and continue to fight COVID-19.
All the authors have agreed to publish the data in your esteemed Journal.
The data and materials have been presented in the main manuscript and can be given upon request.
No competing financial interests exist.
FH (Fuad Hasan) and MA (Md. Arifuzzaman) conceived the idea. FH wrote the manuscript. MA edited the manuscript. All authors read and approved the final manuscript.
This work didn’t receive any funding. Although the authors acknowledges to Department of Genetic Engineering & Biotechnology, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh.
Ethical approval was not required.
Citation: Arifuzzaman. Md,Hasan F(2020) Severe Acute Respiratory Syndrome – Coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19): An overview of Structure, Clinical Features, Diagnosis and Treatment. Cell Dev Biol 2020; 9:214. doi:10.24105/2168-9296.2020.9.214
Received: 03-Nov-2020 Accepted: 18-Nov-2020 Published: 26-Nov-2020 , DOI: 10.35248/2168-9296.20.9.214
Copyright: © 2020 Arifuzzaman. Md, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.