Advanced Techniques in Biology & Medicine
Open Access
ISSN: 2379-1764
ISSN: 2379-1764
Research Article - (2022)Volume 10, Issue 5
According to the new 2016 World Health Organization (WHO) classification of cancers of the Central Nervous System (CNS), Glioblastoma Multiforme (GBM) is the most frequent malignant tumour of the CNS. GBM contains a wide range of genetic and epigenetic changes, resulting in a large number of mutation subgroups, some of which have been shown to play a role in independent patient survival and treatment response. Despite advances in GBM treatment, patients who are diagnosed with these tumours often have a poor prognosis and a low quality of life as the disease progresses. The single-cell RNA high-throughput sequencing processed data for glioma cancer stem cells was taken from GEO-NCBI and was analyzed to find out the underlying expression differences in a single Glioma Neural Stem Cell (GSCs) and Normal Neural Stem Cell (NSCs). We have performed bioinformatics analysis on the transcriptional profile of 134 samples which consisted of 75 GSCs and 59 NSCs obtained from the NCBI bio project (PRJNA546254). An exploratory analysis was performed which showed significant gene expression variation patterns between GBM tumour and normal stem cell. Subsequently, Deseq2 differential gene expression analysis identified 383 differentially expressed genes between GSCs and NSCs [padj. value <0.05, log2 fold change (>=+/-1.5)]. This study reveals genes like LOX, LOX1, COL6A2, COL8A1, COL3A1, LUM, TGFB1, LAMA2, POSTN, MFAP5, MFAP2, FBN2, FLRT2 and HTRA1 as the key features contributing to the disturbed processes of Extracellular matrix organization and extra-cellular structural organization pathways in glioma. Conclusively, the results presented here reveal new insight into the progression of GBM and the identification of novel genes involved in gliomagenesis, which could be looked into further for achieving therapeutic targets.
Glioblastoma multiforme; Single cell RNA-sequencing; Cancer stem cells; Bioinformatics
GBM: Glioblastoma Multiforme; GSCs: Glioma Neural Stem Cells; NSCs: Normal Neural Stem Cells; CSCs: Cancer Stem Cells; PCA: Principal Component Analysis; ECM: Extra-Cellular Matrix.
Gliomas are one of the most common and malignant brain tumours [1]. They can occur anywhere in the central nervous system but primarily occurs in the brain, spinal cord and the glial tissues [1]. However, it is still unknown whether gliomas arise from normal glial cells, glial or neural progenitors, stem cells, or other cell types. Histopathology has traditionally been used to diagnose and classify gliomas. Glial tumours were classified as astrocytic tumours, oligodendroglial tumours, oligoastrocytic tumours, ependymal tumours, neuronal (gangliomas) and mixed neuronal-glial tumours by the World Health Organization (WHO) in 2007 [2]. These includes grade I tumours like pilocytic astrocytomas, pleomorphic xanthoastrocytomas, and subependymal giant cell astrocytomas, as well as more common infiltrating gliomas like grade II oligodendrogliomas and astrocytomas, and grade III anaplastic oligodendrogliomas, anaplastic oligoastrocytomas, anaplastic oligodendroma and Glioblastoma (GBM) [2].
Glioblastoma is a fatal, aggressive brain tumour [3]. The most common and malignant primary brain tumour in adults is glioblastoma. Necrosis and endothelial proliferation are the histopathologic markers that define grade IV, the highest grade in the World Health Organization (WHO) classification of brain tumours. Glioblastomas that are developed from the previously diagnosed WHO grade II or grade III gliomas, are classically referred to as "secondary glioblastoma” [4].
Both in the kreb cycle and in the cytoplasm, the Isocitrate Dehydrogenase (IDH) family of enzymes catalyses the conversion of isocitate to alpha-ketoglutarate while also converting Nicotinamide Adenine Dinucleotide Phosphate (NADP+) to reduced NADP+(NADPH) [5]. In a number of malignancies, somatic mutations in genes encoding two isoforms of Isocitrate Dehydrogenase (IDH1 and IDH2) are found. Although other substitutions at these or surrounding positions are seen, the great majority of mutations are caused by an amino-acid change at position 132 in IDH1 from arginine to histidine (R132H) and at position 172 in IDH2 from arginine to lysine (R172K) [6,7]. In gliomas, all pathogenic mutations occur in the sub-state recognition site, which drastically alters the enzyme active sites, resulting in a neomorphic shift in IDH function. Proteins produced by IDH gene mutations convert alpha-ketoglutarate to the probable oncometabolite 2-Hydroxyglutarate (2HG) [8].
GBM is found in around 80% of malignant gliomas (DeAngelis, 2001; Thakkar et al., 2014). Patients with GBM have a dismal prognosis and, if left untreated, they expire quickly. The majority of patients die within two years of diagnosis, and the overall survival duration is less than a year (Laws et al., 2003; Mirimanoff et al., 2006; Reardon and Wen, 2006; Wen and Kesari, 2008; Rock et al., 2012; Hanif et al., 2017).
While the morphologies of GBM tumours may be similar or overlap, variations in tumour growth and molecular pathways necessitate diagnostic beyond histological profiling [9]. The requisite excision or biopsy can be troublesome due to the location and delicate nature of brain tissue, making histological profiling a task in and of itself. Non-invasive diagnostics that can better identify and discriminate GBM tumours will clearly speed up non-resection therapy techniques, enhancing patient survival [10].
Malignant tumors such as GBM have highly diversified phenotypic and molecular characteristics at both intratumour as well as intertumour levels [11]. Interlesion heterogeneity also known as intertumour heterogeneity refers to the differences in the tumour profiles and characteristics found between different patients [11]. Intralesion heterogeneity also known as Intratumor heterogeneity refers to tumor cell populations (with different molecular and phenotypical profiles) within the same tumor specimen or patient [11]. Oncogenic mutations cause cancer, which is often characterized as a hereditary disease [12]. Intratumoral heterogeneity has long been linked to genetic variability inside cancer cell populations, in a similar gene-centric paradigm. Recent research reveals, however, that a tumour is heterogeneous in practically every apparent phenotypic attribute as a result of non-genetic sources of variability as well as genetic factors [13].
Heterogeneity in tumours is linked to a poor prognosis and outcome [11-13]. Intratumor heterogeneity is regarded to be one of the most important predictors of therapeutic resistance and treatment failure, as well as one of the main causes of poor overall survival in cancer patients with metastatic illness [14]. Different layers of complexity exist in tumour heterogeneity. Individual patients, lesions, and cell populations should be thoroughly characterized at various intervals because cancer is a heterogeneous dynamic target [15]. Tumor heterogeneity has posed a significant barrier in matching patients with the proper medication at the right time, making precision medicine aims difficult to achieve [16].
Cancer Stem Cells (CSCs) (also referred to as tumor-initiating cells) are a source of functional cellular heterogeneity in tumors. Genetic heterogeneity resulting from DNA sequence variation and/or whole chromosomal and focal Copy Number Variations (CNVs) is another source of intra-tumor heterogeneity. Aneuploid or chromosomally unstable populations of tumour cells with aberrant numbers of chromosomes produce whole chromosome CNVs [17]. Aneuploidy is a stable state in which aneuploid cells in a tumour have the same defective karyotype, and it is common in cancer, with aneuploidy being seen in over 90% of solid tumours. Furthermore, many aneuploid tumour cells have Chromosomal Instability (CIN). CIN is characterized by a high incidence of chromosome mis-segregation that results in random chromosome losses and gains [18].
Many efforts have been made by researchers and oncologists to understand the origin of GBM, but the reason still remains unclear. Due to this, the etiology of GBM remains unknown. Out of the many hypotheses, two of them suggest the origin of GBM. One of them suggests that GBM arises from Cancer Stem Cells (CSCs) that possess the ability to self-renew, differentiate and encourage tumor formation. The second theory suggests that GBM progresses due to intertumor heterogeneity [19]. This point’s towards an important area of research for identifying the intertumor differences in the cancer stem cells populations in order to achieve therapeutic targets.
The main goal of this study was to identify the underlying gene expression changes in a single tumour cell owing to intra-tumor heterogeneity. In the present study we investigate the impact of focal Copy Number Variation (CNV) due to chromosomal instability on gene expression profiles by analyzing the transcriptomes of a glioblastoma Cancer Stem Cell (CSC) line, GliNS2 CSCs, that is chromosomally unstable and a control Normal Neural Stem Cell (NSC) line, CB660 NSCs. Besides finding gene expression profile differences intra-tumourly, identification of affected biological pathways would help further in the identification of molecular mechanisms which encourage the rate of molecular hetereogeneicity.
Hence, in the present study, we investigate the transcriptomic profiles of 75 Glioma Neural Stem Cells samples (GSC) and 59 Normal Neural Stem Cells (NSC) samples to understand the gene expression differences among them. After the identification of significant differentially expressed genes among the same tumour sample, we scrutinized the most affected biological pathways which revealed important results into the neuronal processes like axonal growth cone dynamics, extracellular matrix organization, synaptogenesis and progenitor dynamics as the transformations happen from GSCs to GBM or NSCs to GBM. Besides, the results presented here reveal new insight into the progression of GBM due to molecular heterogenecity and the identification of novel genes involved in gliomagenesis and intra-tumour differences.
Software’s used in this study
Server T-bio Info
Website: https://server.t-bio.info/
The Principal component analysis, H-clust and heat map plots was done using this cloud based server.
Metaboanalyst
Website: https://www.metaboanalyst.ca/
Metaboanalyst was used to generate PCA plots using the associated data set of this study. The T-test and volcano plots were also generated using metaboanalyst.
Enrichr pathway analysis
Website: https://maayanlab.cloud/Enrichr/
The gene ontology analysis was performed using the enrichr knowledge database.
Data sets
For this study, the process of single-cell RNA high-throughput sequencing processed data for glioma cancer stem cells was taken from NCBI GEO [GSE132172]. The dataset of this project was generated by Zhao Y et al., (2019) and was published as a bio project on NCBI with accession number (PRJNA546254). This dataset consisted of RNA-Seq data retrieved from CB660 normal neural stem cell lines and GliNS2 glioblastoma stem cell lines. A total of 134 samples were present on the associated SRA run selector, 59 samples of Normal Neural Stem Cells (NSCs), and 75 samples of Glioma Stem Cells (GSCs) were selected and downloaded as an SRA Run. Table 1 represents the same number and the sample size per group.
| Disease state | Number of samples |
|---|---|
| Glioblastoma | 75 |
| Normal neural stem cells | 59 |
Table 1: Tabular representation of sample sizes and groups.
Data pre-processing: The high throughput sequencing processed data was quantile normalized signal data. The processed data had gene symbols. The soft family files had other meta-data information if required.
Down-stream analysis
Principle component analysis: To understand the patterns of gene expression differences among the different genes from the same GBM tumour, comparative data analysis was performed using the Principal Component Analysis (PCA) module integrated on the T-bio info server (https://server.t-bio.info/). PCA is a dimensional reduction technique that is applied to larger data sets, in order to visualize the variation between samples in a particular data set [20]. PCA was performed between these 2 conditions namely: (a) Glioma Neural Stem Cells (GSCs) and (b) Normal Neural Stem Cells (NSCs). Eventually, the PCA plots were generated using metaboanalyst (https://www.metaboanalyst.ca/).
Differential gene expression analysis: The differential gene expression analysis was performed using the Deseq2 [21] tool on the metaboanalyst platform to derive significantly differentially expressed genes between GSCs and NSCs samples. DESeq2 is a tool based method for differential analysis of count data that uses shrinkage estimation for dispersions and fold changes to improve the stability and interpretability of estimates. This helps to enable a more quantitative analysis focused on the strength rather than the mere presence of differential expression. The significant genes were identified with the threshold of [p.adj value <0.05, Fold change (>= ± 1.5)].
Assessment of discriminatory potential of significant genes: In order to visualize and see the clustering patterns obtained from the potential significant genes, H-clustering [22] and a heat map [23] were formed using the 134 intra-tumour GBM samples (75 GSCs and 59 Normal Neural Stem Cells (NSCs)) with the selected set of significant genes. These significant genes were generated using the Volcano plot tests and T-tests which were run using metaboanalyst software. H-clustering is an important technique of machine learning which is used to group similar data points such that the points in each same group are more similar to each other than the points in the other groups. The groups formed are known as “clusters”. H-clustering (distance: euclidean, linkage: average) was performed to understand the discriminatory potential of significant genes in distinguishing the gene expression patterns between the two cell lines of GSCs and NSCs samples. Later, heat maps were drawn using the T-bio-info server in order to elucidate the pictorial representation of specific up-regulated and down-regulated genes out of the potential significant genes obtained using volcano and T-tests.
Gene enrichment analysis: In order to draw out the biological significance of the differentially expressed genes, a gene set enrichment analysis [24] was performed. Gene Set Enrichment Analysis (GSEA) is a powerful analytical method to interpret gene expression data [25]. GSEA helps one find the genes involved in molecular, biological and cellular pathways. Moreover, GSEA was performed using enrichr platform [26], which is an easy-to-use intuitive enrichment analysis web-based tool providing various types of visualization summaries of collective functions of gene lists.
In the present study we analyzed, the transcriptomes of a glioblastoma Cancer Stem Cell (CSC) line, GliNS2 CSCs, that is chromosomally unstable and a control normal, diploid Neural Stem Cell (NSC) line, CB660 NSCs.
Comparative data analysis
The data was further analyzed using the principle component analysis method [27], on the software metaboanalyst. Figure 1 represents the variations between GBM tumour (GSCs) and Normal Neural Stem Cell (NSCs) gene samples. Moreover the Figure 2, below represents a 3D PCA Plot and Figure 3 represents PCA pipeline on T-bio Infor server.
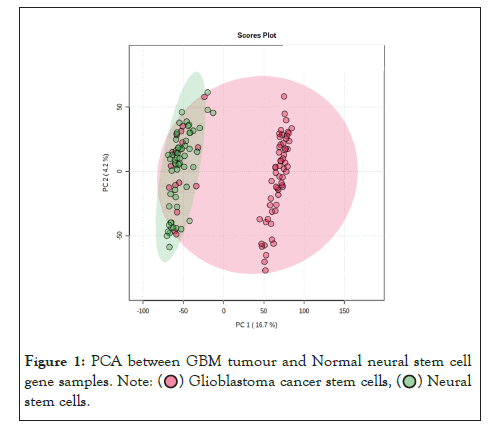
Figure 1: PCA between GBM tumour and Normal neural stem cell
gene samples. stem cells.
stem cells.
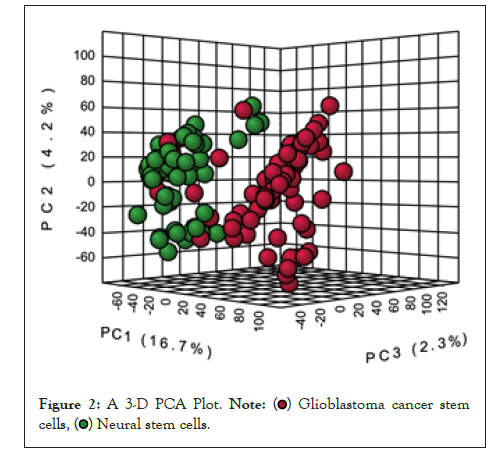
Figure 2: A 3-D PCA Plot.
 .
.
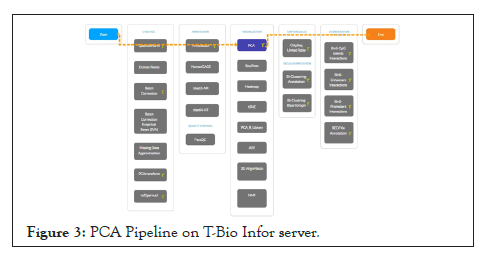
Figure 3: PCA Pipeline on T-Bio Infor server.
Downstream analysis
It is evident from the PCA results in Figure 1 that the variation between the genes of glioma neural stem cells samples and normal neural stem cells samples is maximum. This is observable from the two different clusters formed in PCA 3D Plot and the % differences as well. To further analyze the data we carried out a comprehensive comparative analysis between glioma neural stem cells samples and normal neural stem cell samples. Followed by that, in downstream analysis, we performed differential gene expression analysis between the genes glioma tumour cell sample and normal neural stem cell sample. Also, when the glioma tumour stem cell sample and normal neural stem cell sample was examined in a specific scatter plot [26], it was obvious that there was an improvement in principal components, and the two groups were separated from each other which showed clear distinction. The observable clear distinction between these two groups was enough to lead us further for investigation the tumour’s transcriptomic profiles.
Differential gene expression analysis
The differential gene expression analysis [28] between the genes of Glioma Neural Stem Cells (GSCs) and Normal Neural Stem Cells (NSCs) samples scrutinized 383 significantly differentially expressed genes between GSCs and NSCs [padj. value <0.05, log2 fold change (>=+/-1.5)]. Among them, 109 genes were found to be significantly upregulated (p.adj value <0.05, Fold change >=+1.5) genes and 653 genes were found to be significantly downregulated (p.adj value <0.05, Fold change <=-1.5) in GSCs in comparison to NSCs. A volcano-plot for the differentially expressed genes can be seen in Figure 4.
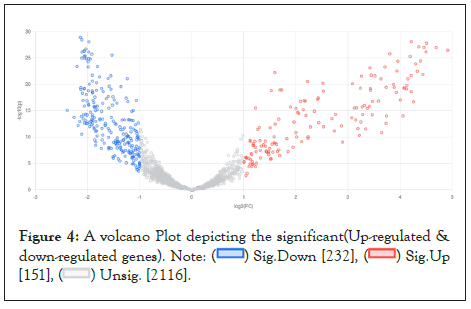
Figure 4:A volcano Plot depicting the significant(Up-regulated &
down-regulated genes).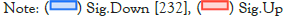
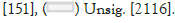 .
.
Clustering and heat map revealed variations among glioma neural stem cells and normal neural stem cells samples
Hierarchical clustering [29] was performed to visualize intra-tumour gene expression variations between the two groups namely GSCs and NSCs tumour. The main aim of hierarchical clustering was to understand if the significantly differentiated genes are capable of forming clusters based on their gene expression profiles. The clustering results show clear distinct clusters formed between GSCs and NSCs samples as shown in Figure 5. The heat map [30] representing the gene expression patterns between GSCs samples and NSCs samples is shown in Figure 6. The heat map indicates specific up-regulated and down regulated genes.
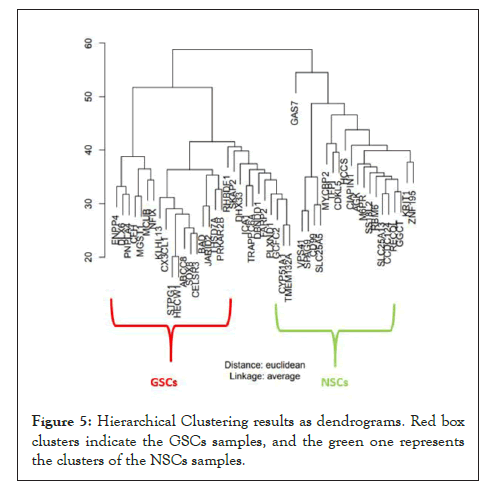
Figure 5: Hierarchical Clustering results as dendrograms. Red box clusters indicate the GSCs samples, and the green one represents the clusters of the NSCs samples.
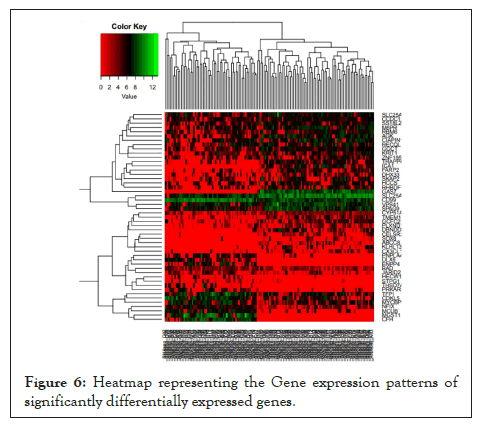
Figure 6: Heatmap representing the Gene expression patterns of significantly differentially expressed genes.
Pathway enrichment analysis
To understand the involvement of the differentially expressed down-regulated and up-regulated significant genes, pathway analysis [31] was performed using the enrichr [32,33] knowledge database. Figure 7 depicts what biological, molecular and cellular pathways are affected due to the up-regulated genes. Figure 8 depicts what biological, molecular and cellular pathways are affected due to the down-regulated genes.
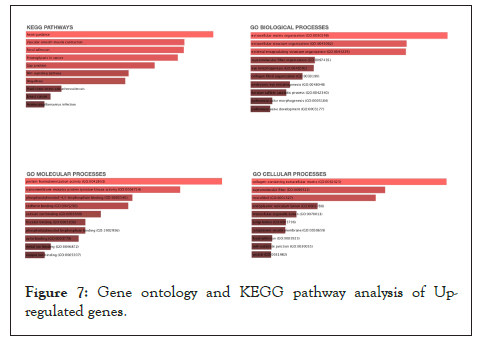
Figure 7: Gene ontology and KEGG pathway analysis of Upregulated genes.
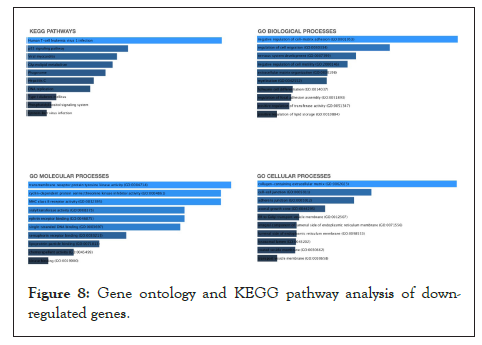
Figure 8: Gene ontology and KEGG pathway analysis of downregulated genes.
The top hits for the major pathways affected due to the up-regulated genes included the axon-guidance pathway (KEGG Pathway analysis), extracellular matrix organization (GO biological pathway analysis), Protein homodimerization activity pathway (GO molecular pathway analysis) and collagen containing extra-cellular matrix pathway (GO cellular pathway analysis).
Similarly, the top hits for the major pathways affected due to the down-regulated genes included the human T-cell leukemia type-1 infection pathway (KEGG pathway analysis), negative regulation of cell matrix pathway (GO biological pathway analysis), P-transmembrane receptor protein tyrosine kinase activity pathway (GO molecular pathway analysis) and collagen containing extra-cellular matrix pathway (GO cellular pathway analysis).
Here on, we down-streamed our analysis, and focused on disturbed extra-cellular matrix organization pathways due to the up-regulation of specific genes. Some of the major genes involved in the progression of disturbed extra-cellular matrix organization pathway are LOX, LOX1, COL6A2, COL8A1, COL3A1, LUM, TGFB1, LAMA2, POSTN, MFAP5, MFAP2, FBN2, FLRT2 and HTRA1.
The origin of glioma and progression of gliomagenesis still remains unknown [34]. With no known specific etiological feature addressed to yet, this necessitates more investigation into the involvement of glioma tumours. We aimed to establish a strong connection between glioma tumour formation and the underlying gene expression alterations in this study using the single cell RNA-Seq analytic approach on the transcriptome profiles of genes within glioma tumor cell and normal neural stem cell. In our comparative analysis, where we performed the principle component analysis, we observed clear clusters formed between GSCs and NSCs tumour cell line sample. This supports the idea that the difference between glioma neural stem cells and normal neural stem cells is due to differences in gene expression among the same tumour at the transcriptomics level. Further studies with larger number of sample sizes would reveal the changes occurring exactly during this transformation at the gene level at the intra-tumour region. Furthermore, based on the differential gene expression analysis using the Deseq-2 tool on the metaboanalyst software between the Glioma Neural Stem Cell tumour (GSCs) and Normal Neural Stem Cell (NSCs) samples we scrutinized 383 significantly differentially expressed genes between GSCs and NSCs [padj. value <0.05, log2 fold change (>=+/-1.5)]. Among them, 109 genes were found to be significantly up-regulated (p.adj value <0.05, Fold change >=+1.5) genes and 653 genes were found to be significantly down-regulated (p.adj value <0.05, Fold change <=-1.5) in GSCs in comparison to NSCs. Followed by that, in order to get the visual representation of the data analysis results we plotted the dendrograms, heat map, and hierarchical clustering which revealed the significance of significantly expressed genes among the GBM tumour and normal stem cell sample. A heat map generated from these significant genes showed that some of the genes were down-regulated in GSCs tumor cell whereas some of them were up-regulated in normal neural stem cell, highlighting differential expression of genes within a single cancer cell population.
Gene ontology analysis performed using enrichr based on significant gene sets showed obvious involvement of genes in pathways like axon growth curve pathway, molecular nervous system development pathway, amyloid beta-binding pathways, some of the amino acids metabolism pathway, axon guidance, extracellular matrix organization, collagen binding pathways, and receptor-ligand binding pathways. Furthermore, one of the most affected pathways due to the up-regulated genes as per our biological processes pathway analysis was the extracellular matrix organization pathway.
The Extracellular Matrix (ECM) is a non-cellular meshwork of cross-linked macromolecules that constitute a dynamic, supra-molecular framework. It delivers physical and chemical cues that drive cancer progression and spread [35]. The Extracellular Matrix (ECM), which is a primary structural component of the tumour microenvironment, is a highly dynamic structure, and there is growing evidence that ECM proteins provide a physical and metabolic habitat for CSCs. A disruption in the balance between ECM synthesis and secretion, as well as altered expression of matrix-remodeling enzymes, causes aberrant ECM dynamics in cancer [36]. However, studies suggest that the tumor-derived ECM has a different biochemical composition and is stiffer than normal ECM.
Sequestered growth factors, ECM biomechanics, and ultrastructural organisation, among the other biochemical and biomechanical signals present in the ECM, are recognized by cells and transformed into downstream cellular responses. These downstream cellular responses work together to slow or stop the progression of cancer. Disrupted turnover of ECM components, as well as aberrant or absent post-translational modification, are among the alterations seen in many illnesses, including cancer. Furthermore, the ECM is a highly ordered structure, and its functional qualities are dependent on ECM component assembly precision. Subtle alterations in these components' stoichiometry may have biological repercussions that affect tissue function. A Cancer-Associated Fibroblast (CAFs) is key stromal cells in the tumour microenvironment that can be trained and/or recruited by tumour released substances [36]. CAFs' ability to manufacture and alter ECM components has a significant impact on tumour growth. Understanding the heterotypic interactions between tumour cells, the ECM, and CAFs in the tumour microenvironment can provide insight into the mechanisms that support tumour growth and metastasis [37].
All this suggests, that ECM has major role in cell migration, cell proliferation, cancer stemness, tissue homeostasis, and collagen secretion. Taking the upper hand, ECM stops the cancer progression while secretion of various proteins such as collagen binding proteins, laminins, heparin family proteins, and integrin’s. However in our study, up-regulation of the extra cellular matrix organization pathway suggests the over activation of this biological pathway. Over activation of any biological processes leads to an enhanced activity of that pathway. Here in the case of up-regulation of extra cellular matrix pathway, it secretes a higher amount of proteins, collagen and other growth factors. Collagen can help cancer cells grow and migrate, but new research has found that it can also impact the function and phenotype of tumor-infiltrating immune cells such Tumor-Associated Macrophages (TAMs) and T cells [38]. Hence over-secretion of collagen allows more cancer stem cells to be transformed to cancerous malignancies and also in cancer tumour cell proliferation and metastasis. The lack of collagen has also been linked to malignancies growing more quickly, according to research. Hence secretion of Collagen binding proteins by ECM in right required amounts imparts immune function of preventing the tumour cell proliferation and re-growth. However Under or over secretion of Collagen due to disturbed ECM pathways provides a way for tumour cell proliferation and other cancerous metastatic pathways.
To overcome this problem and impart therapeutic resistance towards the up-regulation of Extra cellular matrix pathway organization we can target the genes discovered in our study such as LOX, LOX1, COL6A2, COL8A1, COL3A1, LUM, TGFB1, LAMA2, POSTN, MFAP5, MFAP2, FBN2, FLRT2 and HTRA1. Targeting these genes would help us combat the over-activation of the extra cellular matrix organization pathway. This would provide a helping hand towards controlling the GBM tumour cell proliferation.
ECM components in a tumour microenvironment may nevertheless pose a number of problems that could jeopardize an otherwise successful treatment programme. To begin with, ECM proteins have been proven to operate as a physical barrier to medicine delivery to cancer cells. Second, ECM proteins can de-differentiate non-CSCs into CSCs, making it more difficult to eliminate all CSCs. Finally, because ECM influences immune cell recruitment, dysregulated ECM components may obstruct prospective immunotherapeutic techniques. Finally, the ECM is a complex and dynamic system: Different ECM molecules are expressed at different times and in different tissues and different isoforms of the same molecule might play conflicting roles in cancer stemness depending on the circumstances. Given these concerns, future research should focus on elucidating the role of ECM components in cancer stemness in order to develop medicines that effectively eliminate all CSCs.
In future studies, may be accessing the precise role of Collagen protein coding genes in relation with Glioblastoma multiforme cancer could help us achieve therapeutic targets for GBM. However limited number of samples was a limitation of this study, which could be overcome with larger sample size. Apart from this, analyzing the role of genes like LOX, LOX1, COL6A2, COL8A1, COL3A1, LUM, TGFB1, LAMA2, POSTN, MFAP5, MFAP2, FBN2, FLRT2 and HTRA1 which contribute to the significantly up-regulated disturbing process of the extra cellular matrix organization pathway in humans, could help us achieve therapeutic targets for treating this fatal brain tumour.
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
[Crossref][Google Scholar][PubMed].
Citation: Vaja R (2022) Single-cell RNA Sequencing of Glioblastoma Cancer Stem Cell Lines Reveals that Chromosomal Instability Leads to Disturbed Extra-cellular Matrix Organization. Adv Tech Biol Med. 10:359.
Received: 06-Jun-2022, Manuscript No. ATBM-22-17767; Editor assigned: 09-Jun-2022, Pre QC No. ATBM-22-17767 (PQ); Reviewed: 23-Jun-2022, QC No. ATBM-22-17767; Revised: 30-Jun-2022, Manuscript No. ATBM-22-17767 (R); Published: 07-Jul-2022 , DOI: 10.35248/2379-1764.22.10.359
Copyright: © 2022 Vaja R. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.