Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2008) Volume 1, Issue 3
The discovery of short interfering RNA has admitted the development of facile regulated methods for disruption of gene expression. However, this method continues to grow in popularity, designing effective siRNA can be demanding. So we have developed siRNA Scanner, a siRNA selection program that automatically selects Small Interfering RNA from the given RNA sequences. siRNA Scanner uses a fuzzy logic-based system to calculate siRNA qualities. This program is fully built in Practical Extract Report Language (PERL 5.8.8.6 Build 820) and accessible in a command line interface. siRNA Scanner's high performance, minimal user interaction, and its fast algorithm, make this program useful for selecting Small Interfering RNA for gene expression studies.
Keywords: Fuzzy logic, siRNA, Design tool
RNAi was initially observed in petunias, when Napoli et al ., (1990) discovered that introducing a pigment-producing gene suppresses expression of both the introduced gene and the homologous endogenous gene, a phenomenon they called "cosuppression". RNA interference (RNAi) is the process of mRNA degradation that is induced by doublestranded RNA in a sequence-specific manner (Fire et al., 1998; Hannon , 2002). In all RNA silencing pathways, double-stranded RNA (dsRNA) is processed to a small RNA which is assembled with RISC into a silencing complex that specifically represses expression or function of a target gene or genomic region by cleaving the corresponding mRNA (McManus et al., 2002). However, introduction of long dsRNA into mammalian systems is more problematic. This is due to the fact that in most mammalian cells dsRNA, that is more than 40 nt, (Frank Buchholz, unpublished data) induces a nonspecific interferon response, leading to the general shutdown of transcription and/or cell death (Williams, 1997;Stark et al ., 1998). In mammalian system the most standard protocols for RNAi therefore use chemically synthesized siRNAs of 19-22 base pair length for gene silencing. Yet this approach is limited by the fact that different sequences within a gene have dramatically varied inhibitory abilities (Holen et al., 2002). In essence, a large number of different synthetic siRNAs have to be screened for their efficacy at knocking down the gene of interest, which is a laborious and costly task.
Several siRNA design tool exists Ambion siRNA Target Finder (http://www.ambion.com/techlib/misc/ siRNA_finder.html), Jack Lin's siRNA Sequence Finder (http://www.sinc.sunysb.edu/Stu/shilin/rnai.html), siDESIGN Center (http://www.dharmacon.com/sidesign), siRNA Target Finder (https://www.genscript.com/ssl-bin/ app/rnai), Imgenex sirna Designer (http://imgenex.com/sirna_tool.php), EMBOSS sirna (http://bioweb.pasteur.fr/seqanal/interfaces/sirna.html), IDT RNAi Design (SciTools) (http://www.idtdna.com/Scitools/Applications/ RNAi/ RNAi.aspx), BLOCK-iT RNAi Designer (https:// rnaidesigner.invitrogen.com/rnaiexpress), siSearch (http://sonnhammer.cgb.ki.se/siSearch/siSearch_1.7.html), SiMAX (http://www.mwg-biotech.com/html/s_synthetic_acids/s_sirna_design.shtml, BIOPREDsi (http://www.biopredsi.org/), Promega siRNA Target Designer (http://www.promega.com/siRNADesigner/program/), QIAGEN siRNA Design Tool (http://www1.qiagen.com/ Products/GeneSilencing/CustomsiRNA/siRNADesigner. Aspx), SDS/MPI (http://i.cs.hku.hk/~siRNAa/software/ siRNA.php), WI siRNA Selection Program (http://jura.wi.mit.edu/bioc/siRNAext/ ), which use a set of common criteria (e.g. GC%, Tm) for selecting the siRNA candidate in a specific target region selected by the user or in predefined region. Here, we describe a new tool for the designing small interference RNA based on various efficient criteria from the literature and these criteria are calculated for every siRNA candidate over the entire target sequence without masking. Using efficient fuzzy logic syste m, we assigned a quality grade to each of the candidate. To make the application as widely available as possible, the tool is programmed in PERL, which is multiplatform compatible.
siRNA Scanner for the design of functional siRNA’s includes the rules based on the work recently published by different authors (Holen et al., 2002; Reynolds et al ., 2004; and Wuming et al., 2006), which have proven to be more efficient than the consensus rules accepted to date. The novelty of this tool is that it is the only tool till date that applies Fuzzy logic to select the efficient candidate for siRNA interference.
Physical Parameters for Fuzzy Construction
Direct Sequence features (Eight Features)
For each position in the sense strand of 19-mer siRNA sequence, 8 features were defined based on whether or not the nucleotide at the position is an adenine (A), a cytosine (C), a guanine (G), or a uracil (U), respectively (Holen et al ., 2002). Eight features are listed below:
| Feature 1 | 2nd ucleotide | = | A | ||||
| Feature 2 | 4nd nucleotide | = | C | ||||
| Feature 3 | 6nd nucleotide | ≠ | C | ||||
| Feature 4 | 7nd nucleotide | ≠ | U | ||||
| Feature 5 | 9nd nucleotide | = | C | ||||
| Feature 6 | 17nd nucleotide | = | A | ||||
| Feature 7 | 18nd nucleotide | ≠ | C | ||||
| Feature 8 | 19nd nucleotide | = | (A/U) | ||||
Percentage of GC Content
The GC content of the siRNA duplex is an obvious candidate for a parameter that might correlate with siRNA functionality. Too high GC content may slow down duplex unwinding by the putative helicase entatively associated with the RISC complex (Reynolds et al ., 2004) and might also be associated with a prohibitive secondary structure of the target mRNA. Too low GC content, on the other hand, may reduce the efficiency of target mRNA recognition and hybridization. The strongest correlation was observed for the GC range 31.6–57.9% (Reynolds et al ., 2004) and this range is considered as optimum in our fuzzy based rule.
Thermodynamics
Three quality grades were defined based on whether or not the melting temperature (Tm) falls into following ranges 0°C – <20°C, >20°C–<60°C, and >60°C.
No Occurrences of Four or More Identical Nucleotides in a Row
This feature implies that a siRNA sequence have no internal repeated sequence of length >= 4.
No Occurrences of G/C Stretches of Length 7 or Longer
This feature implies that a siRNA sequence have no GC stretch of length > 7 or longer.
At least 3(A/U)'s in the Seven Nucleotide at the 3’ end
Based on the number of A/U in the seven nucleotide at the 3’-end, the feature is categorized into Least, At least, High. Least falls under <3 A/U, At least >= 3 A/U<5, and High >=6.
Computational Methods
Fuzzy Logic
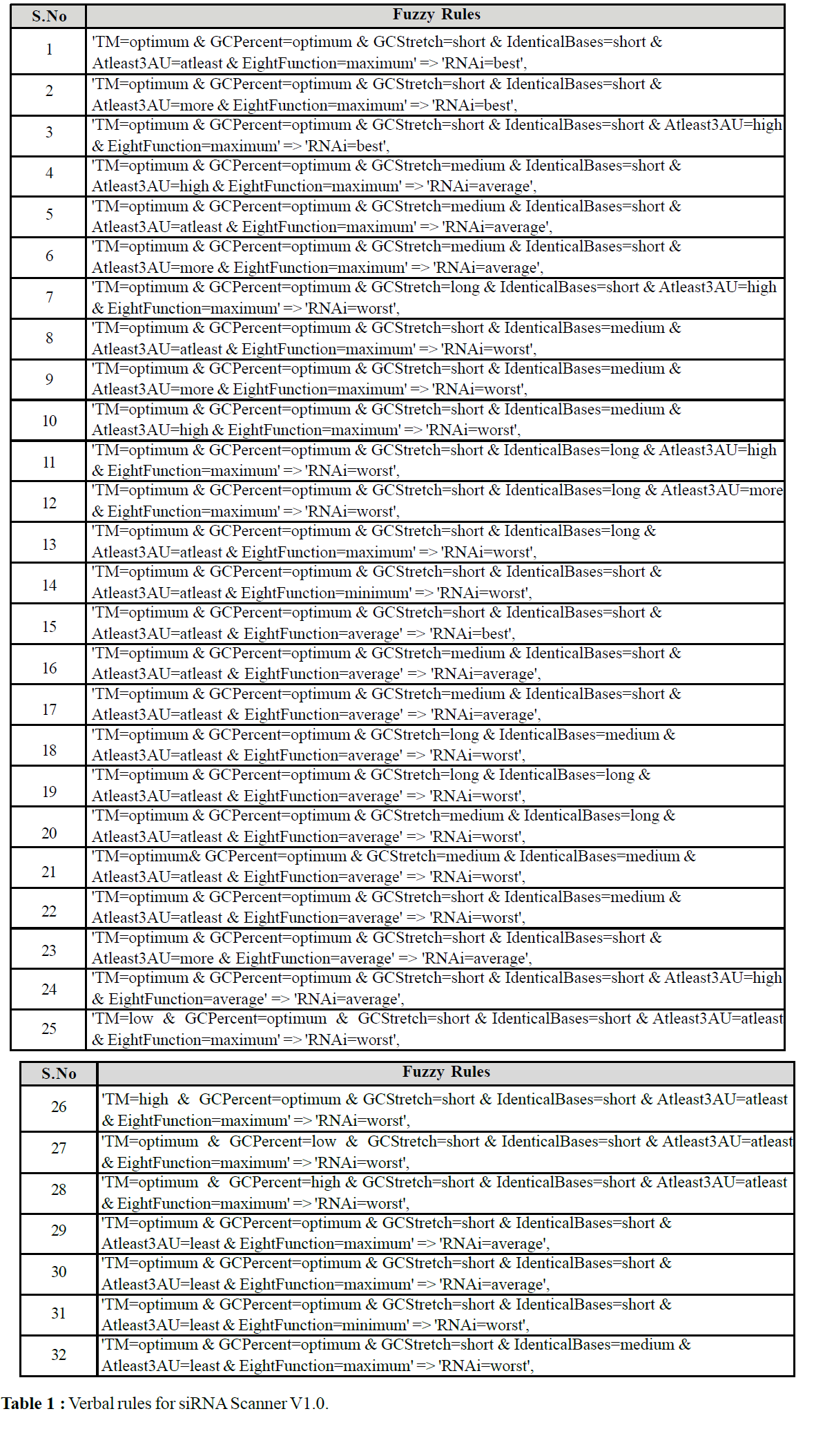
For the purpose of assigning relative weights to the parameters Simulink’s Fuzzy logic Tool box (MATLAB®) was used. This tool takes a list of relevant parameters as input and their ranges are partitioned into categories according to specifications by the user. For example, the GC% content was categorized into Low, Optimum, High and Very High. These categories may be overlapping and we used specific membership functions to describe partial assignment of a value to a category. Table 1 lists a set of the rules used in this application. The fuzzy logic system designs a mathematical model that maps the input parameters to output parameters in accordance with the user’s qualitative description. It is thus, not necessary for a user to design a mathematical description by himself. Instead, this description is generated by the fuzzy logic system based on only a verbal description of the relevant rules.
Data Storage
siRNA Scanner does not reject any siRNA candidate before having calculated all relevant parameters for each possible 19mer. For efficient handling of the huge amount of data generated during a siRNA candidate search we store all relevant information in a tree. This structure can be built up in a time proportional to the number of positions in the sequence using the on-line-construction algorithm of Ukkonen (Ukkonen, 1995).
Programming Language
siRNA Scanner V1.0 is entirely coded in PERL (Practical Extract Report Language) Version 5.8.8 Build 820. Three different modules namely, AI::FuzzyInference and Math::Fortran was downloaded from the CPAN archive and used in this application.
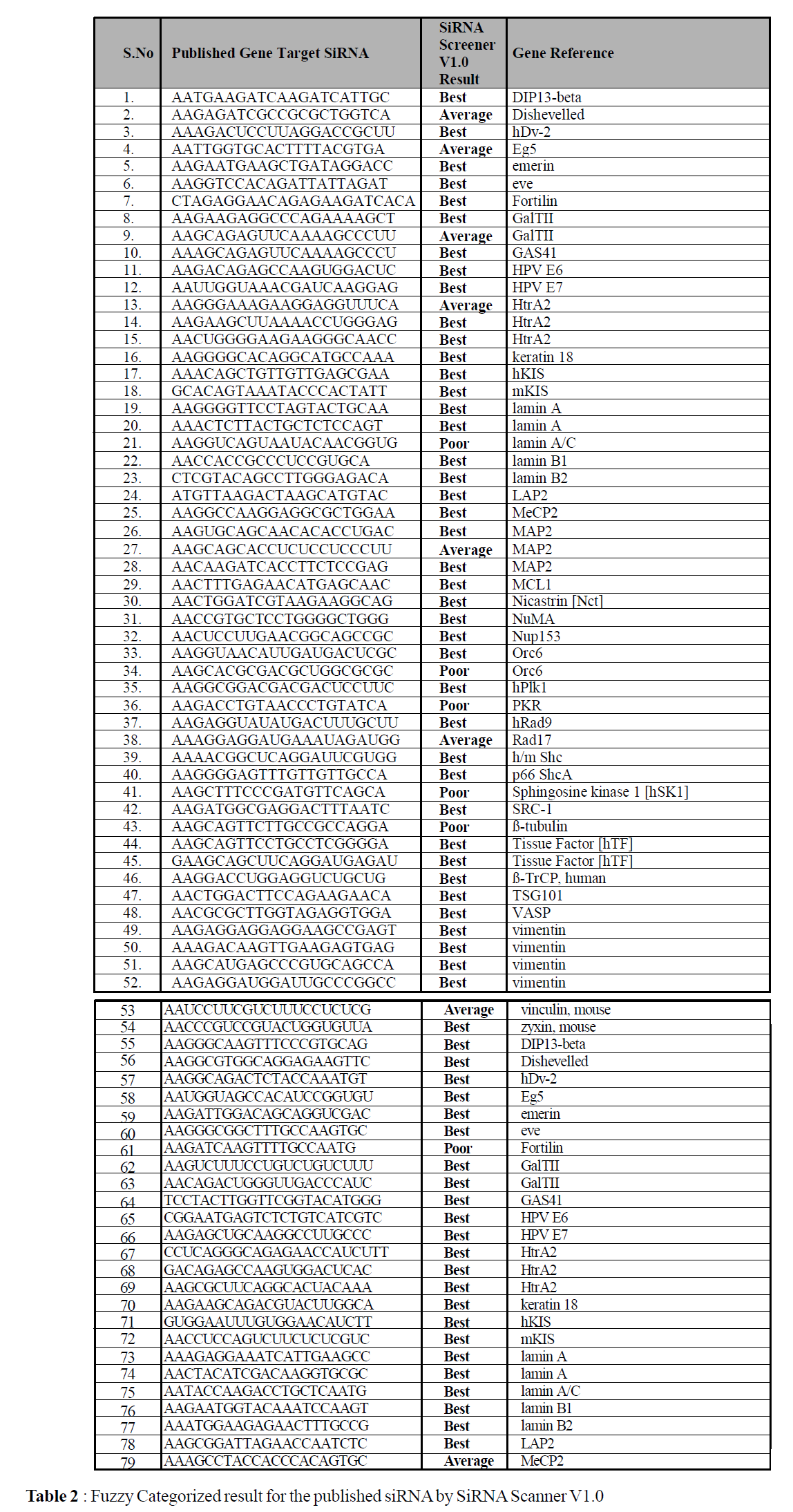
The procedure for siRNA selection has been implemented in PERL as a stand-alone tool. The use of this program is simple and one needs to input the template nucleotide sequence file alone as input. The file format is plain text. Initially the program splits all possible candidates for the calculation, then in the second phase it calculates the fuzzy rules, assigns weight based on the membership function and finally defuzzify and categorize based on the rule framed (Table 1) each candidate as either Best or Average or Poor. Once the siRNA Scanner completes the task, it query the user for five different options for the filtering the result. It includes (a) Best SiRNAs Alone (b) Average SiRNAs Alone (c) Best & Average SiRNAs Alone (d) Poor SiRNAs Alone (e) All Possible SiRNAs (Fig.1). The result produced from the tool is a hypertext file with user defined name and the result are presented as table (Fig .2).
A test set of 79 genes, which already contains the reported active SiRNA’s from literature, have been tested with the siRNA Scanner to analyze its success rate. Each gene sequences are collected from the NCBI website and fed into our tool for the prediction of possible efficient siRNA candidate. Out of 79 genes, for 69 gene set our tool predicted the published siRNA candidate as "Best", which is around 87.34% of the total input, for 6 gene set our tool reported the published siRNA candidate as “Average”, which is around 7.25% of the total input; summing up siRNA Scanner predicted the published siRNA as the possible candidate with the efficiency of 94.93%, that is 75 out of 79 siRNA candidate (Table 2), which is published as efficient gene silencer. Rest of the 4 candidate is predicted as "Poor" by our tool. Evaluation of test set shows an efficiency rate of 94.93% for siRNA candidate prediction. siRNA Scanner’s high performance, minimal user interaction, and its fast algorithm, make this program useful for selecting Small Interfering RNA for gene expression studies.

Three main objectives have been addressed in this work that makes siRNA Scanner a tool with efficient feature compared with other similar programs: speedy calculation; efficient prediction based on fuzzy logic; and considers all possible candidate without discarding any 19mers. In contrast to other siRNA design software, siRNA Scanner evaluates each possible siRNA candidate. The amount of computational effort here is comparatively best than other siRNA design program. siRNA Scanner is fast in calculating optimal siRNA’s. It checks for all possible candidates from the given template sequence. Calculation time depends linearly on the length of sequence used for checking. On average, a siRNA calculation considering all possible candidates on a target sequence of length 5000 bases pairs takes ~ 21 seconds on an IBM Machine with Pentium 4 Processor. siRNA Scanner’s high performance makes it feasible to efficiently search siRNA on large templates sequence.