Journal of Physical Chemistry & Biophysics
Open Access
ISSN: 2161-0398
ISSN: 2161-0398
Research Article - (2022)Volume 12, Issue 4
The methodology of Specific Peptides (SP) has been introduced within the context of enzymes. It is based on unsupervised Machine Learning (ML) tool for motif extraction, followed by supervised annotation of motifs. In the case of enzymes, the classifier is the Enzyme Classification (EC) number. Here we restudy this problem, and demonstrate that we reach precision of 0.965 and recall of 0.891 on presently available protein sequences. Moreover, applying our methodology to query proteins is much faster than deep learning methods used for the same purpose.
We also apply this method to two other protein groups, G Protein Coupling Receptors (GPCR) and zinc finger proteins, find their corresponding SPs, and provide the code for searching any protein sequence for its classification under any such family. Some proteins which have annotations belonging to two of the three systems are being discussed. Our methodology can be applied to any protein group in order to find their corresponding SPs and provide the code for searching any protein sequence for its classification under any such family.
Specific Peptides (SP); Enzymes; GPCR; Zinc fingers
Genes were perceived well before they have been determined to exist on chromosomes. In hindsight, it seems quite a surprise to find that they are just stretches of nucleotides within much larger sequences of DNA, often also interspersed by noncoding sections (Introns). The identity of genes comes to life after being transcribed into RNA molecules, and translated into proteins, the important components of the machinery of living cells. Proteins are molecular chains of amino acids. They are being studied by investigating the linear composition of amino-acid sequences, or their folding structures, or their functional properties, as revealed by their interactions with other molecules. In this paper, we discuss a different perspective of their structures, resulting from amino acid motifs, which are observed to be common to many proteins having the same function, belonging to homolog genes of different species.
We follow the methodology developed and tested [1-4], pointing out the existence of Specific Peptides (SPs) which are motifs of length ≥ 7 amino acids, occurring on enzymes only. Although motifs of shorter lengths may also be useful [5], we limit ourselves to length ≥ 7 in order to obtain higher precision. We reanalyze all enzymes using the updated Enzyme Classification (EC) labelling, employed by Swiss-Prot [4]. This analysis demonstrates the high predictive power of enzymatic SPs which will be labelled ESPs.
The analysis starts with the motif extraction method MEX [6], which is an unsupervised algorithm finding motifs with high occurrence in a given text. The method has been first developed for ML studies of linguistic texts, and later applied to biological texts, such as amino acid chains in proteins. There exist half a million annotated Swiss-Prot proteins [4], and about half of them are enzymes. Following traditional ML methodology, we use 90% of the enzymes as a positive training set Ptrain, on which we conduct our Motif Extraction (MEX) search. Once the motifs are extracted, we employ supervised labelling providing the motifs with EC labels, according to the EC assignments of proteins on which they occur. Next we test for their occurrence on a negative Ntrain set, containing 90% of all non-enzymatic proteins in the data. Motifs which are found to have hits in Ntrain are discarded, and all the rest are declared to be Enzymatic Specific Peptides (ESP). The prediction accuracy is finally tested on the remaining 10% of the data, Ptest and Ntest.
The methodology is a slightly improved version of the older analysis [1-3], which did not include the negative training mode. It is carried out on a larger and updated list of proteins, employing an updated version of the enzyme classification list (which includes a novel 7th category). We then expand our analysis to proteins which have other functional assignments: GPCR proteins, including both Olfactory Receptors (OR) and the multitude of non-OR proteins (which we label as NOR), continuing with the important set of all zinc finger proteins. There exist some overlaps between the different sets which we point out and demonstrate exhibiting the power of the corresponding novel SP sets: GSPs (containing distinctive OR and NOR ones) and ZSPs recognizing zinc finger domains in proteins.
SPs should be considered within the context in which they were derived. They are not supposed to annotate a free peptide, but only the motif appearing within a protein sequence. Still, as such, they can help identifying and annotating novel proteins, and may turn out to be very useful for artificial protein engineering [7] and for medical research and development [8].
Motif Extraction (MEX)
The MEX algorithm was developed within a linguistic study [6], and later applied to strings of biological alphabets (such as amino acids and nucleotides). The basic idea is to study certain texts (e.g., Protein sequences) and extract motifs, i.e., certain substrings, which appear many times in the text without any change. When such motifs are found, they are tested for their specificity to certain texts (Protein families). When considering proteins, we note that a motif of length ≥ 7 has a very low probability to randomly occur multiple times in the data. Hence it is a sign of homology, indicating loci of structure and/or biological function which can be associated with such specific Peptides.
Building the list of ESPs
Half of the annotated proteins in Swiss-Prot are enzymes. Dealing with a list of more than 200 K entries, we divided the enzymes training set into batches grouped by joint level 2 assignments, and batches of enzymes with single level 1 assignments. We restricted our MEX search to motifs of length ≥ 7 amino acids [3]. The analysis led to 307,989 motifs. All motifs were then annotated after collecting the information of the IDs of enzymes hit by a particular motif (i.e., occurring in full on the amino acid chain of the enzyme) and how many times was a particular enzyme hit by a particular motif.
The EC number description, indicating both class and level, can be viewed as an inverted tree with a maximum depth of 4. For every motif, we map the EC numbers of the enzymes it hits on the training set onto a single EC tree. Starting from level 4 and moving upwards, we search the first level which is a unique descendent of a higher level. The EC number of this unique descendant is assigned to the motif.
In order to remove motifs which may occur also on non- enzymatic proteins, we search for hits of all motifs on the non- enzymatic Ntrain set. Such motifs are removed from the list of specific peptides. Thus, to summarize, a motif of length ≥ 7 amino acids is labeled as an Enzyme Specific Peptide (ESP), presented as Set 2 in Table 1, if:
| SP set | TP | FP | FN | TN | Precision | Recall |
|---|---|---|---|---|---|---|
| 1 | 22722 | 2664 | 2479 | 27160 | 0.895 | 0.902 |
| 2 | 22283 | 806 | 2716 | 28910 | 0.965 | 0.891 |
| 3 | 20821 | 66 | 4469 | 29369 | 0.997 | 0.823 |
Note: We use conventional definitions of Precision=TP/(TP+FP) and Recall=TP/(TP+FN). The sizes of Ptest and Ntest are 25,309 and 29,416 correspondingly. The FP events in sets 2 and 3 include mismatched EC assignments from Ptest as well as SP hits on proteins of Ntest. Thus the 806 in the case of Set 2 includes 300 from Ptest and 506 from Ntest. See further explanation in results.
Table 1: Classification of enzymes according to 3 sets of SPs.
• It hits (i.e., appears in full on the amino acid chain of) enzymes belonging to only a single EC classification of Ptrain.
• It does not hit any protein in Ntrain.
This procedure leads to the reduction of the set of motifs to 286,755 specific peptides which we label as ESPs. They are provided as a json file in our github entry which also includes a Python program (SPs.py) to search for ESPs within a protein's string of amino acids and generate an EC prediction for the queried protein [9].
Lists of GSPs and ZSPs
The numbers of GPCR and ZF proteins are in the thousands, two orders of magnitudes smaller than the number of enzymes. Hence we use all of them for training purposes, and check later on for specificity to particular protein families. We also run sanity checks for their occurrence on other types of proteins. We note and discuss the existence of enzymatic properties of some particular GPCRs, and the occurrence of enzymatic regions on ZF proteins.
The Python program (SPs.py) provided in [9], can be used to query amino sequences for GPCR or ZF predictions using the “-dSPs” parameter pointing to the appropriate json file: ESPs.json for enzymatic predictions, dZFs.json for zinc finger predictions or dGPCR.json for GPCR predictions.
Enzyme specific peptides
The Swiss-Prot entry (version 2021_01) contains 564,227 proteins of many species [4]. In order to enable training and testing procedure we divided randomly the enzymes which had a single EC annotation into two sets: 227,488 were designated to a positive training set (Ptrain) and 25,309 enzymes were designated to a positive test set (Ptest) . The single EC annotation constraint has been introduced in order to allow for a unique EC assignment in the automatic supervised labelling procedure. In parallel we also constructed non-enzymatic negative training and test sets, Ntrain and Ntest, containing 264,739 and 29,416 proteins correspondingly. Ntrain serves to discard motifs which are not specific to enzymes.
Using the Enzyme Classification (EC) nomenclature, enzymes are classified into seven classes, EC1 to EC7, and within each EC class they are grouped into a hierarchy of four levels. Some are classified just into the first level, numbered by the class, some at levels 2 or 3, but most at level 4, which is often associated with homologs of the same gene in different species. Proteins which have enzymatic regions belonging to two different EC classes were discarded from the training set, but the different regions can be discovered on the same protein using ESP searches.
In order to test the usefulness of ESPs in predicting the EC labelling of a protein, we ran it on the test sets Ptest and Ntest. We ask whether the ESP prediction is consistent with the EC number of the enzyme. An SP hit on P-test is regarded as True Positive (TP) if the Swiss-Prot EC assignment of the enzyme appears on the EC tree of the SP, otherwise it is regarded as False Positive (FP). If no SP hits an enzyme, it is labelled as False Negative (FN). If an SP hits a protein in Ntest, the latter is declared as False Positive (FP). If no SP hits a protein in Ntest, it is regarded as True Negative (TN).
In Table 1 we present statistics which correspond to three SP sets. We restricted our MEX search to motifs of length ≥ 7 amino acids [4]. This leads to the existence of 297,404 SP candidates, based on Ptrain only. We label this set as Set 1. Running all this set on Ntrain we find hits by 10,649 motifs, which we discard henceforth. The result is Set 2 containing 286,755 specific peptides, which becomes our standard set of ESPs. Note that the Ntrain pruning of motifs had a relatively small effect: Only 3.58% of motifs have been discarded. In other words, even restricting ourselves to positive data only, such as Set 1, MEX provides trustable results. The reason must be that long substrings of amino acids have a very small probability of being incidental. Set 3 is extracted from Set 2 by excluding predictions due to a single SP hit of length 7 or 8. This can be stated as an additional constraint, demanding the SP coverage of the protein (meaning the number of its amino acids which are hit by ESPs) to be at least 9.
The 3 digits' accuracy quoted for precision and recall is due to the large numbers of Ptest and Ntest. Running the same statistics on 5 different random fractions of 50% of the test sets leads to the same average results, with standard deviation less than 10-4.
The difference between sets 2 and 3 of Table 1 represents predictions due to a single SP hit, of length 7 or 8, on a protein. There are 4404 such cases out of the total of 54,725 test proteins. The precision and recall of such single hits are 0.66 and 0.46 accordingly. Precision rises above 0.9 for all single SP hits with length ≥ 9.
Set 2 is chosen as our standard set of ESPs. Its details are provided in the Supplementary Material. They are also provided as a json file in our github entry [9], which includes the code for searching a protein for the occurrence of such ESPs.
G Protein Coupling Receptors (GPCR)
G Protein Coupling Receptors (GPCR) play dominant roles in olfaction, vision and many other cellular functions. They serve as cell surface receptors, and all have seven transmembrane sections. Olfactory Receptors (OR) was studied by using motifs of length ≥ 5 derived by the MEX methodology. Gottlieb A, et al. [5], have demonstrated how the resulting motifs can be employed to sketch an evolutionary tree of species, and have provided a web-service for OR protein assignment on the basis of these motifs. We limit ourselves to motifs of length ≥ 7 to assure higher precision, and extend our analysis to all GPCRs listed by Swiss-Prot.
The total number of OR proteins in Swiss-Prot is 562, including 469 listed for human. The number of Non-OR (NOR) proteins is 2481, with only 148 in human. On the ORs we find 367 motifs with length ≥ 7, while the NOR proteins lead to 3710motifs. The two different motif classes are exclusive, i.e., we do not have motifs of one class hitting a protein in the other class. The larger number of NOR motifs is explained by the fact that they belong to many different protein families serving a large number of functional modalities. These families are listed in Table 2. Motifs which are specific to a given family are regarded as SPs and listed as such in Table 2. Other motifs, which are common to more than one family of proteins, are counted separately in the column labelled “motifs”. The list of GPCR SPs, which we refer to as GSPs, is divided into OR and NOR groups, and is presented in the supplementary material and in our github file [9].
| # | Function | # Proteins | # SPs | # Motifs |
|---|---|---|---|---|
| 1 | 5-hydroxytryptamine receptor | 93 | 98 | 151 |
| 2 | Adhesion G protein-coupled receptor | 49 | 92 | 134 |
| 3 | Alpha adrenergic receptor | 52 | 62 | 92 |
| 4 | Angiotensin II receptor | 22 | 22 | 31 |
| 5 | Beta adrenergic receptor | 49 | 78 | 116 |
| 6 | Blue-sensitive opsin - Green-sensitive opsin - Rhodopsin | 156 | 160 | 269 |
| 7 | Cadherin EGF LAG seven-pass G-type receptor | 9 | 30 | 62 |
| 8 | Chemokine-like receptor | 146 | 108 | 177 |
| 9 | Dopamine receptor | 43 | 49 | 76 |
| 10 | Frizzled | 53 | 117 | 119 |
| 11 | G protein-coupled receptor kinase | 12 | 19 | 40 |
| 12 | Galanin receptor type | 11 | 3 | 10 |
| 13 | Gamma-aminobutyric acid type B receptor subunit | 4 | 1 | 5 |
| 14 | Gastric inhibitory polypeptide receptor | 7 | 3 | 10 |
| 15 | Gastrin/cholecystokinin type B receptor | 10 | 9 | 14 |
| 16 | Golgi pH regulator | 9 | 13 | 18 |
| 17 | Gonadotropin-releasing hormone receptor | 17 | 11 | 20 |
| 18 | G-protein coupled bile acid receptor | 5 | 3 | 4 |
| 19 | G-protein coupled receptor | 163 | 160 | 273 |
| 20 | Growth hormone-releasing hormone receptor | 11 | 6 | 13 |
| 21 | Histamine receptor | 20 | 41 | 60 |
| 22 | Hydroxycarboxylic acid receptor | 6 | 2 | 4 |
| 23 | Latrophilin Cirl | 10 | 66 | 83 |
| 24 | Leukotriene B4 receptor | 4 | 2 | 4 |
| 25 | Lutropin-choriogonadotropic hormone receptor | 12 | 13 | 28 |
| 26 | Lysophosphatidic acid receptor | 17 | 11 | 17 |
| 27 | Medium-wave-sensitive opsin | 27 | 50 | 44 |
| 28 | Melanin-concentrating hormone receptor | 6 | 4 | 5 |
| 29 | Melanocortin receptor | 19 | 12 | 21 |
| 30 | Melanocyte-stimulating hormone receptor | 81 | 118 | 146 |
| 31 | Melanopsin | 10 | 9 | 13 |
| 32 | Melatonin-related receptor | 24 | 10 | 23 |
| 33 | Metabotropic glutamate receptor | 45 | 100 | 146 |
| 34 | Muscarinic acetylcholine receptor | 35 | 73 | 108 |
| 35 | Mu-type opioid receptor | 13 | 5 | 25 |
| 36 | N-arachidonyl glycine receptor | 5 | 3 | 4 |
| 37 | Neuromedin receptor- Neuropeptide receptor | 47 | 29 | 50 |
| 38 | N-formyl peptide receptor | 15 | 16 | 24 |
| 39 | Nociceptin receptor | 5 | 6 | 8 |
| 40 | Orexin receptor type | 10 | 17 | 25 |
| 41 | Oxytocin receptor | 13 | 10 | 22 |
| 42 | P2Y purinoceptor | 29 | 23 | 39 |
| 43 | Parathyroid hormone/parathyroid hormone-related peptide receptor | 12 | 16 | 28 |
| 44 | Pituitary adenylate cyclase-activating polypeptide type I receptor | 4 | 1 | 7 |
| 45 | Platelet-activating factor receptor | 8 | 7 | 10 |
| 46 | Prokineticin receptor | 8 | 7 | 11 |
| 47 | Prostaglandin receptor | 29 | 26 | 39 |
| 48 | Proteinase-activated receptor | 18 | 11 | 19 |
| 49 | Proto-oncogene Mas | 5 | 1 | 2 |
| 50 | Relaxin receptor | 7 | 5 | 8 |
| 51 | Serpentine receptor class | 18 | 8 | 14 |
| 52 | Short-wave-sensitive opsin | 11 | 11 | 22 |
| 53 | Smoothened homolog | 4 | 12 | 13 |
| 54 | Somatostatin receptor type | 21 | 13 | 23 |
| 55 | Sphingosine 1-phosphate receptor | 16 | 14 | 19 |
| 56 | Substance-K receptor | 10 | 5 | 19 |
| 57 | Taste receptor member | 49 | 99 | 141 |
| 58 | Thromboxane A2 receptor | 5 | 3 | 4 |
| 59 | Thyrotropin receptor | 14 | 24 | 34 |
| 60 | Trace amine-associated receptor | 44 | 41 | 58 |
| 61 | Urotensin-2 receptor | 5 | 4 | 6 |
| 62 | Vasoactive intestinal polypeptide receptor | 19 | 5 | 16 |
| 63 | Vasopressin receptor | 12 | 14 | 23 |
| 64 | Vomeronasal type-1 receptor | 26 | 22 | 39 |
Note: # SPs refers to motifs which are specific to a single family, while # motifs refers to other motifs occurring on several NOR families and not classified as GSPs.
Table 2: 64 protein families belong to NOR GPCR.
Since the number of proteins used in this study is quite small, especially when compared to all enzymes, we have resorted to positive training only. Next we test the specificity of GSPs by searching their hits on all enzymes. We find 63 hits on three enzyme families, listed below in Table 3, reflecting the fact that these GPCRs serve indeed as enzymes. Thus these proteins carry Other hits of GSPs on enzymes can serve as error indicators. We find only 20 sporadic hits of NOR GSPs on all other enzymes, a data base of over 200000 proteins. Hence we conclude that a false. Positive error of GSPs is negligibly small, of order of 10-4.
| EC classification | NOR classification |
|---|---|
| 2.7.11.14 Rhodopsin kinase | Blue-sensitive opsin, Green- sensitive opsin, Rhodopsin |
| 2.7.11.15 (Beta-adrenergic-receptor) kinase | Beta adrenergic receptor |
| 2.7.11.16 (G-protein-coupled receptor) kinase | G protein-coupled receptor kinase |
Table 3: Three NOR families which belong to three EC numbers.
Zinc finger proteins
Zinc Finger proteins play very special roles in binding to DNA and RNA. They carry one or more Zinc Finger modules which preform the binding. The ZF modules differ from each other in specific loci which determine the identity of the nucleotides to which they couple.
We have analysed 2582 Swiss-Prot ZF proteins and extracted 1487 motifs of length ≥ 7 which are declared to be ZSPs. 786 of all the proteins are human ZF proteins, and they display hits by 1412 of the ZSPs. We have applied only positive MEX searches, due to the small overall number.
Since ZF proteins may contain several ZF domains, we may encounter reappearance of motifs on different locations within the same protein. This is different from our previous studies of EC and GPCR proteins, where mostly inter-protein multiple appearances were responsible for the generation of MEX motifs. Clearly the repetitive appearances of SPs on a given protein reflect the existence of many ZF regions on the same protein. The latter is usually larger than the number of repeats of a single SP, since different SPs may belong to different ZF regions.
To illustrate these phenomena, we display in Table 4 some ZSPs, which have 100 or more hits on all human ZF proteins, and their occurrences on some ZF proteins. It should be realized that SPs of length n can be contained within SPs of length >n, as can be seen in this table. Summary of all ZSPs and their hits on ZF proteins is provided in our github entry and in the supplementary material [9].
| Protein/SP | CEECGKA | GEKPYKCEEC | HKIIHTG | HTGEKPY | HTGEKPYKCE | KCEECGK | PYKCEECGK | RIHTGEK | YKCEECG |
|---|---|---|---|---|---|---|---|---|---|
| A6NK75 | 9 | 7 | 3 | 5 | 5 | 9 | 8 | 2 | 10 |
| A6NN14 | 25 | 14 | 11 | 13 | 13 | 26 | 20 | 1 | 22 |
| A6NNF4 | 10 | 12 | 4 | 11 | 10 | 13 | 11 | 4 | 12 |
| A8MQ14 | 0 | 0 | 0 | 23 | 0 | 0 | 0 | 14 | 0 |
| A8MTY0 | 7 | 6 | 4 | 8 | 5 | 7 | 7 | 3 | 8 |
| A8MXY4 | 18 | 5 | 6 | 5 | 5 | 19 | 14 | 0 | 15 |
| O43345 | 19 | 20 | 1 | 18 | 17 | 28 | 25 | 4 | 27 |
| O75346 | 6 | 5 | 0 | 9 | 5 | 5 | 5 | 3 | 5 |
| O75373 | 7 | 8 | 2 | 6 | 6 | 8 | 6 | 6 | 8 |
| O75437 | 7 | 6 | 4 | 6 | 6 | 10 | 9 | 1 | 11 |
| O95780 | 6 | 4 | 0 | 6 | 4 | 5 | 4 | 4 | 5 |
| P0DKX0 | 11 | 6 | 1 | 4 | 4 | 14 | 7 | 4 | 12 |
| P0DPD5 | 8 | 5 | 1 | 6 | 5 | 9 | 7 | 7 | 9 |
| P17019 | 9 | 7 | 4 | 9 | 7 | 11 | 10 | 1 | 11 |
| P17038 | 14 | 5 | 2 | 7 | 6 | 13 | 11 | 3 | 13 |
| P35789 | 11 | 5 | 0 | 10 | 5 | 9 | 8 | 2 | 9 |
| P52742 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 8 | 0 |
| Q02386 | 0 | 5 | 0 | 13 | 6 | 6 | 6 | 3 | 7 |
| Q03923 | 5 | 5 | 6 | 12 | 6 | 6 | 5 | 2 | 6 |
| Q03924 | 7 | 3 | 1 | 4 | 2 | 6 | 4 | 2 | 6 |
| Q03936 | 9 | 4 | 5 | 6 | 5 | 9 | 8 | 1 | 9 |
| Q03938 | 8 | 3 | 1 | 4 | 2 | 8 | 7 | 5 | 7 |
| Q05481 | 21 | 12 | 6 | 18 | 13 | 21 | 14 | 7 | 21 |
| Q14593 | 5 | 3 | 3 | 6 | 3 | 5 | 5 | 2 | 6 |
| Q5SXM1 | 3 | 4 | 0 | 10 | 4 | 4 | 4 | 10 | 6 |
| Q68DY1 | 9 | 4 | 2 | 5 | 4 | 9 | 8 | 2 | 9 |
| Q6ZN08 | 4 | 7 | 5 | 9 | 7 | 8 | 8 | 3 | 9 |
| Q6ZR52 | 12 | 6 | 2 | 5 | 5 | 11 | 10 | 2 | 11 |
| Q86V71 | 7 | 6 | 0 | 7 | 4 | 6 | 6 | 3 | 6 |
| Q8IW36 | 6 | 3 | 0 | 4 | 3 | 6 | 6 | 4 | 6 |
| Q8IYB9 | 10 | 2 | 1 | 9 | 2 | 6 | 3 | 4 | 6 |
| Q8IYN0 | 6 | 5 | 1 | 6 | 4 | 6 | 5 | 2 | 6 |
| Q8N7Q3 | 11 | 9 | 4 | 8 | 8 | 13 | 12 | 3 | 13 |
| Q8N972 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 3 | 0 |
| Q8TAQ5 | 2 | 2 | 0 | 15 | 2 | 2 | 2 | 11 | 2 |
| Q8TD23 | 9 | 4 | 2 | 5 | 5 | 8 | 8 | 3 | 8 |
| Q8TF20 | 7 | 5 | 0 | 13 | 5 | 7 | 5 | 11 | 7 |
| Q8TF32 | 8 | 5 | 5 | 6 | 5 | 7 | 4 | 3 | 6 |
| Q96IR2 | 0 | 5 | 0 | 17 | 4 | 0 | 0 | 5 | 0 |
| Q96N22 | 9 | 3 | 3 | 8 | 3 | 5 | 4 | 3 | 5 |
| Q96N38 | 9 | 4 | 2 | 5 | 4 | 8 | 4 | 2 | 6 |
| Q96SE7 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 3 | 0 |
| Q9H7R5 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 7 | 0 |
| Q9H8G1 | 4 | 4 | 4 | 7 | 4 | 5 | 4 | 3 | 5 |
| Q9HCG1 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 5 | 0 |
| Q9P255 | 8 | 6 | 3 | 5 | 4 | 8 | 7 | 2 | 9 |
| Q9UII5 | 9 | 6 | 5 | 12 | 7 | 11 | 7 | 3 | 9 |
| Q9Y2Q1 | 9 | 6 | 4 | 6 | 6 | 9 | 8 | 1 | 8 |
| Shown hits | 364 | 246 | 108 | 436 | 230 | 386 | 316 | 187 | 376 |
| Total hits | 473 | 322 | 127 | 2136 | 327 | 509 | 391 | 966 | 477 |
Table 4: Number of hits by different ZSPs, displayed on different human ZF proteins. Large numbers correlate with the fact that many ZF regions can be found on the same protein.
There exist some proteins which act as enzymes and possess zinc fingers. One outstanding example is PRDM9. This protein serves recombination hotspots during meiosis by binding nucleotides with its zinc fingers. The annotations of the human version of this protein are provided [10]. They contain 14 ZF regions. The first starts at location 388 and has length of 24 amino acids. The second starts at 524 and is of length 23, which is also the length of all the following ZFs. In Figure 1 we display the loci of hits by all ZSP and ESP motifs of length ≥ 7 on this protein. All ZF domains have the structure YVCRECxxxxxxxxHQRTHT, where the additional 8 amino acids, replaced by x, vary according to the nucleotide targeted by the ZF domain. To guide the eye we note prevalent occurrence of the structure HQRTHTGEKPYVCRECGRGF which includes the suffix of a previous ZF domain and the prefix of the next ZF domain. Colors and font sizes reflect occurrences of hits by ZSPs and ESPs.
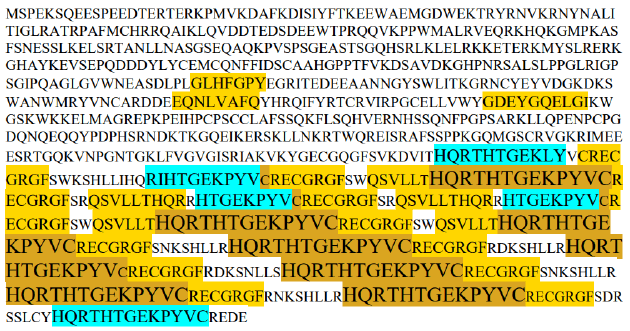
Figure 1:The sequence of PRDM9_HUMAN Histone-lysine Nmethyltransferase (Q9NQV7) and color coded
display of hits by ESPs of EC 2.1.1.43 and ZSPs, which may partially overlap each other. Note: The sequence
of PRDM9_HUMAN Histone-lysine Nmethyltransferase (Q9NQV7) and color coded display of hits by ESPs
of EC 2.1.1.43 and ZSPs, which may partially overlap each other.  is an ESP,
is an ESP,  is a ZSP,
is a ZSP, are overlapping hits by a ZSP and an ESP.
are overlapping hits by a ZSP and an ESP.
Our methodology for predicting enzyme functions is based on Machine Learning (ML) practices. MEX is an unsupervised tool for motif extraction; these motifs are then searched on protein sequences using supervised annotation to classify the results. In the case of enzymes, the classifier is the Enzyme Classification which is defined in terms of seven classes and four levels in each class. This allows us to accurately predict the functions of enzymes, even when partial or incomplete information is available.
ESPs are specific peptides whose presence on the amino acid sequence of the protein indicates its EC number, as well as the tree associated with it.
This methodology was introduced in 2007 [1]. Other ML studies appeared in the meantime, trying to solve the same (or related) problems using various ML tools. Some examples of recent ML methodologies are DeepEC [11], MAHOMES [12], CatFam [13- 15], DETECT [16], ECPred [17], EFICAz2.5 [18], PRIAM [19].
DeepEC employs 3 deep convolutional neural networks and a homology analysis tool to the study of enzyme sequences. When applying it to a test set which uses 201 enzymes they obtained precision=0.92 and recall=0.455 (quoted from Table 2). This is considerably worse than our results in Table 1, which were based on a much larger (25K) test set. Other five ML methods which Ryu JY, et al. [11], compared themselves to, were even worse.
We conducted a comparison of our methodology vs. DeepEC on the same server, against the same file containing 25,309 enzymes from P-Test (Table 5). We used the Aho-Corasick algorithm for efficient search of Specific Peptides within a sequence of amino acids [20]. We found that not only that our Precision and Recall results are better, but our computational speed is 100 times faster than DeepEC’s, with much lower memory utilization and no parallel processing. This is a significant finding, as it demonstrates the potential of our methodology to become the new standard in enzyme classification.
| Precision | Recall | Processing time per sequence (Seconds) |
|
|---|---|---|---|
| DeepEC | 87.50% | 79.70% | 0.05 |
| Specific peptides | 89.50% | 90.20% | 0.0005 |
Table 5: Comparison of DeepEC results to our method using the same test set and the same server.
We have demonstrated the usefulness of our MEX unsupervised methodology in discovering relevant and unique motifs, the Specific Peptides (SPs). Our approach is not limited to enzyme studies. We have shown its flexibility by investigating GPCR and Zinc-finger proteins, leading to a wealth of novel SPs. We provide a documented python code which allows for SP searches of all the functionalities which we have studied. It contains the lists of 2,002 NOR GSPs, 351 OR GSPs and 1,482 ZSPs in addition to the 286,755 ESPs. The lists of all SPs are also provided in the Supplementary Material.
SPs are extracted from persistent homology signals. They may be used for functionality searches in proteins in addition to, or as replacement of, standard alignment searches. Biological roles of ESPs have been demonstrated [2]. SPs may therefore be expected to have functional importance and, as such, should be of interest to medicine and synthetic biology.
Our precision/recall results attest to the usefulness of the MEX unsupervised methodology in discovering relevant and unique motifs, the Specific Peptides (SPs). Our approach is not limited to enzyme studies. We have demonstrated this flexibility by investigating GPCR and Zinc finger proteins, leading to a wealth of novel SPs. We provide a documented python code which allows for SP searches of all the functionalities which we have studied [9]. It contains the lists of 2,002 NOR GSPs, 351 OR GSPs and 1,482 ZSPs in addition to the 286,755 ESPs. The lists of all SPs are also provided in the Supplementary Material. SPs are extracted from persistent homology signals.
They may be used for functionality searches in proteins in addition, rather than as replacement of, standard alignment searches. Biological roles of ESPs have been demonstrated. SPs may therefore be expected to have functional importance and, as such, should be of interest to medicine and synthetic biology.
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[GoogleScholar] [PubMed]
[GoogleScholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
[Crossref] [GoogleScholar] [PubMed]
Citation: Horn D, Weingart U (2022) Specific Peptides Predict Protein Classification. J Phys Chem Biophys. 12:338.
Received: 28-Nov-2022, Manuscript No. JPCB-22-20579; Editor assigned: 30-Nov-2022, Pre QC No. JPCB-22-20579 (PQ); Reviewed: 14-Dec-2022, QC No. JPCB-22-20579; Revised: 21-Dec-2022, Manuscript No. JPCB-22-20579 (R); Published: 28-Dec-2022 , DOI: 10.35248/2161-0398.22.12.338
Copyright: © 2022 Horn D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.