Journal of Physical Chemistry & Biophysics
Open Access
ISSN: 2161-0398
ISSN: 2161-0398
Research Article - (2017) Volume 7, Issue 2
We consider a twisted Peyrard-Bishop-Dauxois (PBD) model and construct the exact analytical solutions, which can describe the propagation of solitary waves by invoking a discrete Jacobian elliptic function method. These solutions include the Jacobian periodic solution as well as bubble solitons. Through the Fourier series approach, we have found that the DNA dynamics is governed by a modified discrete nonlinear Schrodinger (MDNLS) equation. A detailed analysis of the role of the twisted angle in the process of bio energy localization is presented in the form of coherent localized breather modes in a PBD model. A linear stability analysis is performed and we obtain that the stability of the solutions also depends on the twisted angle.
<Keywords: Bioenergy; Jacobian periodic solution; DNA; Biophysicists
Understanding the dynamics of biological processes such as transcription, duplication or DNA translocation by viruses is a challenge for biophysicists. The local opening of the DNA double helix at the transcription start site is a crucial step for the genetic code. This opening is driven by proteins, but the intrinsic fluctuations of DNA itself probably play an important role. The dynamical properties of these bubbles and their relations to biological functions have therefore been the subject of many experimental and theoretical studies. Several models have been used for studying bubble breathing and in attempts to explain these long experimental bubble life-times. i) The Poland- Scheraga model [1] is a one-dimensional (1D) Ising model modified to account for the entropic penalty of creating a closed flexible loop. This term leads to a non-monotonic free energy landscape in which the typical breathing time comes from a Kramer’s process [2-4]. ii) Yomosa’s dynamic plane-base rotator model [5], which involves rotational motion of bases [6,7] and later extended by Zhang [8] by expressing the interstrand interaction through hydrogen bonds in terms of a double-well potential. iii) In the model [9], transverse and longitudinal displacements of the bases were represented in terms of the Toda potential. iv) The Peyrard-Bishop model is a non-linear phonon 1D model where bubbles emerge as soliton-like solutions of undamped Newton’s equations in a Morse potential (possibly with a noise term) [10-12]. This model describes, in a simplified way, the hydrogen bond and has been used successfully in numerous applications such as energy localization [13] or to calculate solitonic speed [14,15]. Experiments proved that the free energy of opening base pairs depends on the identity of the next base pairs; this is due to the stacking interaction between neighboring bases on the same strand [16]. As is well known, the solitons existing in the PB model result from the balanced competition between dispersion and nonlinear effects. In the huge taxonomy of the models for DNA dynamics, the possibility that nonlinear effects might focus the vibrational energy of DNA into localized coherent structures is indeed expressed by considering pulse waves, kinks, or breathers [17]. The impact of a protein interaction on the breather dynamics of DNA by extending the model of Peyrard and Bishop, which is more accurate for the formation of localized oscillations in terms of breathers and bubbles [18]. Since the DNA molecular chain is sequence or site dependent, the strands are flexible and the molecule is helical in shape. Recently, detailed analytical study of the base-pair opening in an inhomogeneous continuum DNA chain in terms of perturbed kink-antikink and bubble solitons, respectively [19,20]. However, the Hamiltonian model proposed so far suffers of a serious short-coming as it does not account for the helicoidal structure and solvent interaction which would have the immediate effect of bringing closer to each other non-consecutive bases along the molecule backbone [21,22]. As a first step to describe helicity in the path integral method, [23,24] recently suggested that one may generalize the stacking Hamiltonian by introducing the angle of rotation between a base pair and the previous one. The twist angle, which increases from one base-pair to the next one, is responsible for the helicoidal structure of the molecule and capture the importance of nonlinear effects on the thermodynamical properties. In B-DNA at room temperature, one turn of the helix hosts about ten base pairs [25]. Accordingly, the equilibrium twist angle, eq=2π/10.4, is expected to provide the energetically most favorable configuration, the stablest one against thermal disruption of the base pair bonds. Also, nonlinear interactions between atoms in DNA can give rise to intrinsically localized breather-like vibration modes [26]. It has been shown that the inclusion of anharmonicities in the study of lattice models can produce qualitatively new effects. In particular, Rosenau and Hyman [27] found solutions of the solitary type without infinite tails, termed solitons with compact support or compactons. These traveling wave solutions have a remarkable property. Interestingly enough, analytic methods have been used to find some analytical solitonic and periodic wave solutions for the DNA molecules.
The exact solutions of nonlinear partial differential equations help us to understand the mechanism of complicated phenomena of the natural systems. Motivated by the above works and applications of excat solutions of DNA molecule, we aim in this work to explicitly present analytical solitary and periodic wave solutions for the twisted (PBD) model with solvent interaction. The paper is organized as follows: in section 2, we briefly present the twisted PBD model of DNA dynamics and then we derive the (MDNLS) equation. Using the Jacobian elliptic function approach, we obtain some exact solutions of the model under study in section 3. In section 4, we study the stability of these solutions. The last section presents some concluding remarks.
Brief presentation of the twisted DNA with solvent interaction
The PBD Hamiltonian for a system of N base pairs assumes the pair mates separation yn (for the nth base pair) with respect to the ground state position as the relevant degree of freedom. The longitudinal base displacements along the molecule backbone are neglected as they are much smaller than the transverse stretching yn. Hence, the model Hamiltonian is essentially one-dimensional. The general form of the PBD Hamiltonian which incorporates, nonlinearities both in the interbase pair interactions and in the coupling between neighboring base along the two strands, helicity and solvent interaction is considered. The Hamiltonian is composed of the following elements.
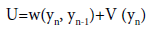 (1)
(1)
where yn is the stretching of the nth base-pair, w(yn, yn-1) is the stacking interaction of the nearest neighbours n and n+1, and V (yn) the interaction of the nth base-pair. In this work, we consider two terms for the base-pair interaction,
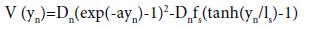 (2)
(2)
The first term is the usual Morse potential which describes the hydrogen bonds of the base-pairs. The second term is a solvent interaction potential on the form presented in Ref. [24] which simulates the formation of hydrogen bonds with the solvent once the base-pair hydrogen bonds are stretched by more than a value ls from their equilibrium values. The solvent interaction potential is a function which varies smoothly from fsD at yn<-ls to zero for yn>ls. For yn
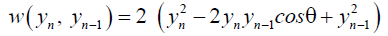 (3)
(3)
where θ is the twist angle between neighboring base-pairs, and one takes K=60 meVA. This is motivated by 3D helicoidal models [29], as well as torsional potentials used in molecular dynamics [28], and is introduced as a way to avoid the divergence of the partition function [30]. For an angle of θ=0 the usual harmonic stack-ing interaction term [31] is obtained and would represent the situation of perfectly parallel neighboring bonds. Evidently, the base-pairs can only de-naturate when the double helix is largely uncoiled and therefore we use a small angle [32,33]. One also consider the anharmonic Hamiltonian [34,35], where the same torsion angle dependence is introduced as in Equation 3 for the stacking interaction,
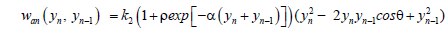 (4)
(4)
ρ and α are the anharmonic stacking parameters which are also assumed independent of the type of base at the n and n-1 sites Thus, generalizing the DPB Hamiltonian to the following expression:
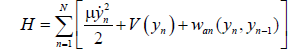 (5)
(5)
K is the harmonic stacking related to μ by K=μν2, ν being the frequency of the phonon mode. The AT and GC base are comparable in size and weight with their effective masses usually taken as μ=300 amu. From this Hamiltonian, an equation governing the local oscillations of DNA nucleotides is derived.
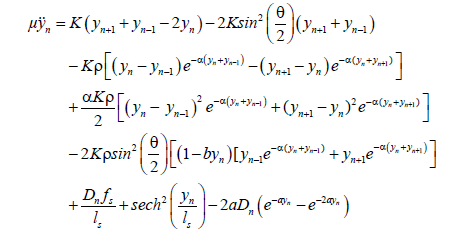 (6)
(6)
In the following, we consider a special collective motion of the pairs in a DNA molecule. The amplitude of the wave packet is big enough, so that the nonlinear effect plays an essential role in DNA molecules. On the other hand, it is still very small compared with the amplitude of the broken base pairs. Therefore, the base pairs in DNA molecule do not oscillate far away from the bottom of the Morse potential well. In this respect, we can assume 0<αyn ? 1, 0
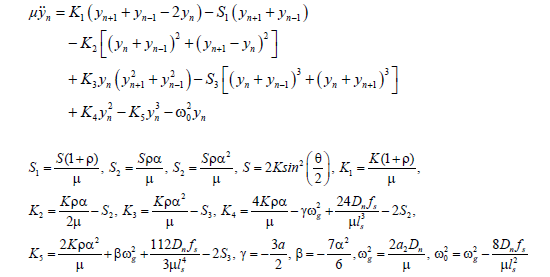 (7)
(7)
Where,
Linearizing Equation (7) yields plane wave solutions with wavenumber k and frequency ω(k) given by the dispersion relation 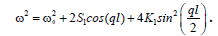 For the nonlinear Equation (7), we search for small amplitude time periodic solutions as
For the nonlinear Equation (7), we search for small amplitude time periodic solutions as
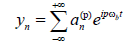 (8)
(8)
where ωb is close to some linear oscillation frequency and the Fourier coefficients are slowly depending on time, 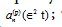 we have defined the smallness implicit parameter
we have defined the smallness implicit parameter 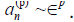 Due to exponential decay of the Fourier
Due to exponential decay of the Fourier
coefficients in p, they must satisfy 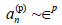 for p>0, while
for p>0, while 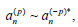 . More-over,
. More-over,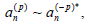 since yn is real. This allows for a slow time dependence of the Fourier coefficients
since yn is real. This allows for a slow time dependence of the Fourier coefficients 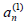 to derive a MDNLS equation for the dominating coefficient
to derive a MDNLS equation for the dominating coefficient 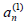 describing the leading-order nonlinear effects. Defining
describing the leading-order nonlinear effects. Defining
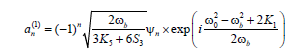 (9)
(9)
the modified DNLS equation of the DNA model reads
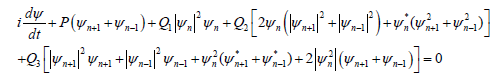 (10)
(10)
where, 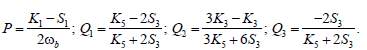
For θ=0, we have Q1=1, Q3=0, 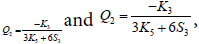 Eq. (10)
Eq. (10)
becomes as the one studied in Ref. [34]. For 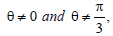 Q2 = 0, Q1>0 and Q3<0, we have here a modified version of the DNLS equation. In the case where,
Q2 = 0, Q1>0 and Q3<0, we have here a modified version of the DNLS equation. In the case where, 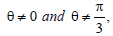 Equation 10 is also a modified DNLS equation.
Equation 10 is also a modified DNLS equation.
Jacobian elliptic solutions for the twisted DNA with solvent Interaction
Seeking exact soliton solutions to nonlinear partial differential equations (PDEs), which describes many complex systems in the field of condensed matter systems, plasma physics, astrophysics, biological systems, etc, is one of the fundamental studies of nonlinear science. Several powerful methods have been proposed to solve differentialdifference equations and to obtain exact solutions to the nonlinear partial differential equation. One can think of the inverse scattering method [36], the tanh method [37], the Jacobian elliptic function method [38], the multilinear variable separation, homogeneous balance method [39], the Backlund transformation method [40] and so on. A uniform method to find all the solutions of nonlinear wave equations does not exist. The periodic wave solutions in terms of the Jacobi elliptic functions for the nonlinear PDEs have attracted considerable interest [41] because of the elegant properties of the elliptic functions. The main idea of this method is to take full advantage of the elliptic equation that Jacobi elliptic functions satisfy and we use the discrete Jacobian elliptic function method to obtain the special Breather-like solutions that govern the transport of energy in DNA chains. At the outset, we make transformations
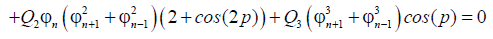 (11)
(11)
replacing ψn in equation (10), and separating the real from the imaginary part we get the following set of equations:
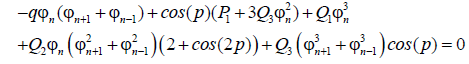 (12)
(12)
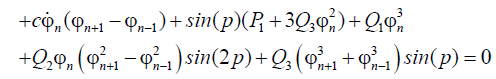 (13)
(13)
With the properties of the Jacobian elliptic function, we use the following series expansion as a solution of Equations (12) and (13) i.e.,
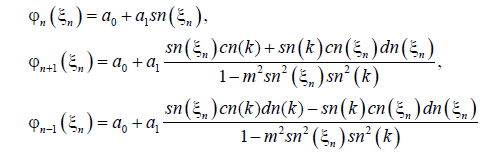 (14)
(14)
Now, we substitute the above equations into Equations (12) and (13), a0, a1, c and q are to be determined. We find after some calculations the following solutions:
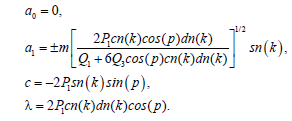 (15)
(15)
and hence the function ?n and ψn are defined as
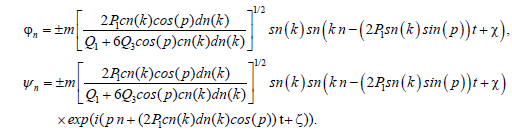 (16)
(16)
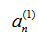 can then be written, taking the + sign:
can then be written, taking the + sign:
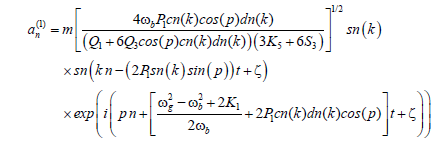 (17)
(17)
The general solution of Equation (4), giving the displacement of the base pairs, i.e., yn, take the form:
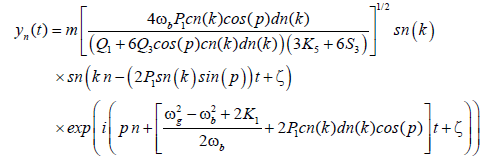 (18)
(18)
Figure 1 presents the profile of the above solution of base pairs for different values of the modulus m of Jacobian elliptic functions. One can see on Figure 1a that as the modulus m of the Jacobian elliptic function increases, the magnitude of the base pairs through the DNA also increases. If the modulus of the Jacobian elliptic function approaches the value 1, one observe on Figure 1b that the solutions of the base pairs are described by the bubble soliton solution. This soliton solution translate the bubbles of transcription observed during the denaturation of the DNA as described in Davydov’s theory. Figure 2 shows the profile of the solution as a function of twisted angle for different values of the modulus m of Jacobian elliptic functions. Through these Figures, we note that the twisted angle has an impact on solutions obtained, pointing the role of this parameter. The twisted angle is important in the process of denaturation of the DNA. Indeed, in a realistic DNA dynamics, the topological constraints related to the helicoidal structure of the molecule could not be ignored. Activation or repression effects caused by conformational changes and, in particular, prevention of transcription due to a large positive excess in twist, are largely investigated in biology. During all processes in which the DNA base-pairs open, a local open, a local unwinding of the helix follows for topological reasons. Consequently, a local extra-twist accumulates at the ends of the bubble and induce a long range elastic stress.
Figure 1: Profile of the solution: (a) the asymptotic evolution of the solutions according to the values of m,m=0.3 (blue), 0.5 (green) and 0.6 (black); (b) the asymptotic evolution of the solution towards the bubble soliton, m=0.99 (blue), 0.9998 (red) and 1 (yellow) and for θ=0.25π, ωb=1, ρ=2, D=0.04 eV, α=0.35 Å-1, K=60 meV.Å-2, fs=0.3, and ls=3 Å.
Figure 2: Profile of the solution according to the values of θ for n=10: (a) the asymptotic evolution of the solutions according to the values of m,m=0.3 (blue), 0.5 (green) and 0.6 (black); (b) the asymptotic evolution of the solution towards the bubble soliton, m=0.9998 (blue), 0.99999 (red) and 1 (black), and for ωb=1, ρ=2 D=0.04 eV, α=0.35 Å, K=60 meV.Å-2, fs=0.3, and ls=3 Å.
Figure 3 depicts the propagation of the Jacobi solution Figure 3a and the bubble soliton Figure 3b through the DNA molecule. One can remark that as the waves move along the molecule, their magnitude increases.
Stability analysis
In this section, we investigate the stability of the above solution. In doing so, we introduce the following expansion:
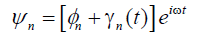 (19)
(19)
Substituting Equation (19) into the MDNLS equation, we find that the linearized equation satisfied by γn(t) is given by
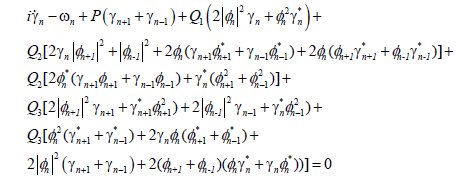 (20)
(20)
Expanding γn(t) in real and imaginary parts: γ0(t) = un(t)+ivn(t) the linearized equations can be written as:
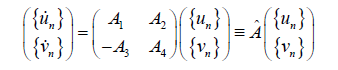 (21)
(21)
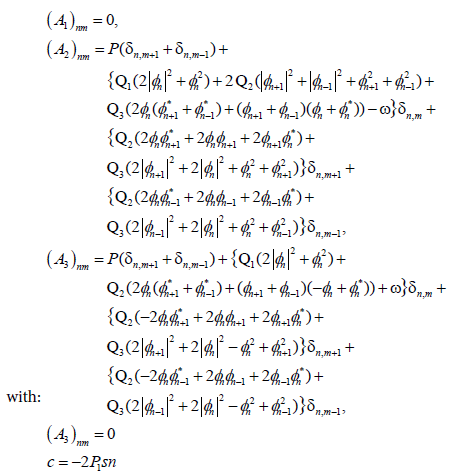 (22)
(22)
by using the periodic boundary conditions, we study the stability of exact solution Jacobi and bubble soliton (see Figure 4) through the eigen value spectrum of the matrix A. The stationary soliton solution is linearly stable if and only if the matrix has all its eigenvalues on the imaginary axis (λ=λr+iλi).
We observe through Figure 4 that for certain value of θ, solutions are stable. For the value of m=1, and θ=0, or θ=0.75π, the spectra plane (λi, λr) of the Jacobi solutions shows all the eigenvalues to be in the imaginary axis, so one can say that the Jacobi solutions are stable. On the other hand, we see on Figure 5 that the eigenvalues leave the imaginary axis for certain values of θ (θ=0.35π and θ=0.55π); thus, the bubble soliton here is unstable for the parameters used. In fact, have already pointed out the fact that bubbles and dark solitons are always unstable [36,42,43], whereas pulse and discrete breather are always stable [39]. Bubble solitons of the discrete cubic-quintic NLS equation are stable due to the presence of the quantic nonlinearity. In this work, we show through Figures 4a and 4c that for a certain value of the twisted angle (θ=0 and θ ≥ 0.56π), the bubble soliton become stable. If one introduces a stochastic white noise in the system, we find that the Jacobi solution is modified during its propagation through the molecule as depicted in Figure 6. The noise effect destroys the coherence of the initial solution [44].
We have considered the twisted DNA with solvent, looking for exact soliton solutions through the Jacobian elliptic function. Using Fourier transform, we shown that the twisted PBD model can be modelled through the MDNLS, with the parameters P, Q1, Q2 and Q3 depending on the twisted angle θ. The solutions found here include the Jacobi periodic solution as well as the bubble soliton depending on the value of the modulus of the Jacobi function. We shown that these solutions depend on the twisted angle θ. The stability of the obtained solutions have been checked. We have found that Jacobi solutions are stable for certain value of twisted angle (θ=0 and θ ≥ 0.6π), and the bubble solitons are also stable for θ=0 and θ ≥ 0.56π.
AM acknowledges the Abdus Salam International Center for Theoretical Physics (ICTP) Trieste-Italy for financial support through the Associateship program.