Journal of Theoretical & Computational Science
Open Access
ISSN: 2376-130X
ISSN: 2376-130X
Research Article - (2017) Volume 4, Issue 1
Statistical optimization was done for L-methionine production by Corynebacterium glutamicum X300 using Response Surface Methodology emphasizing Central Composite Design with different variables. Maximum production of L-methionine (52.1 mg/ml) was obtained with 72 h of incubation.
<Keywords: Statistical; Optimization; L-methionine; Response surface methodology; Central composite design
Statistical methods are widely used for fermentation optimization, because they reduce the total number of experiments required and provide a good understanding of the interactions among different factors on the outcome of the fermentation [1]. Response Surface Methodology (RSM) is a group of mathematical and statistical techniques for the optimization of multiple variables and levels in a minimum acceptable number of experimental trials [2]. Taguchi’s method has gained worldwide acceptance in the optimization of fermentation processes [1].
Adinarayana and Ellaiah used RSM for the optimization of the medium components for the production of alkaline protease by a Bacillus sp [3]. Balusu et al. used RSM for the optimization of medium components for ethanol production from cellulosic biomass by Clostridium thermocellum SS19 [4]. Shih and Shen applied RSM to optimize the production of poly €-lysine by Streptomyces albulus IFO 14147 [5]. Nelofer et al. optimized the process variables for L-lysine production by Corynebacterium glutamicum K [6]. Pandey and Banik optimized different physical parameters for alkaline phosphate production by Bacillus licheniformis using RSM [7].
RSM is advantageous over conventional methods as it requires less number of experiments and its suitability for multiple variable experiments and search for common relationship between different variables towards finding the most suitable conditions for the production of the desired metabolites [8-12]. This methodology could employ to optimize different physic-chemical conditions for L-methionine production.
Thus, the aim of this present investigation was to optimize different production conditions of L-methionine by the mutant Corynebacterium glutamicum X300 controlling different physical and nutritional parameters using RSM.
Microorganism
Corynebacterium glutamicum X300 developed by induced mutantion and protoplast fusion was used throughout the study [13].
Composition of growth medium
Glucose, 20 g; (NH4)2SO4; 1.6 g; NaCl, 2.5 g; MgSO4.7H2O, 0.25 g; MnSO4.4H2O, 0.1 g; K2HPO4, 1 g; KH2PO4, 1 g; H2O, 1 L and agar, 2% as a solidifying agent [14].
Growth conditions
The fermentation was carried out using medium volume, 30 ml; initial pH 7; shaker speed, 200 rpm; the age of inoculum, 48 h; cell density, 3 × 108 cells/ml and temperature, 30°C [14].
Composition of basal salt medium for L-methionine production
L methionine production was initially carried out (before optimization) using the following basal salt medium: glucose, 60 g; (NH4)2SO4, 1.5 g; K2HPO4, 1.4 g; MgSO4·7H2O, 0.9 g; FeSO4·7H2O, 0.01 g; biotin, 60 μg and H2O, 1 L [14].
Analysis of L-methionine
Descending paper chromatography was employed for detecting L-methionine in the broth and was run for 18h on Whatman No.1 chromatography paper. Solvent system contained: n-butanol: acetic acid: water (2:1:1). The spot was visualized by spraying a solution of 0.2% ninhydrin in acetone and quantitative estimation of L-methionine in the suspension was done using a colorimetric method [14].
Confirmatory test for L-methionine
Quantitative determination of L-methionine in the fermentation medium without purification was done following the method as described by Greenstein and Wintz. 1 ml of 5(N) NaOH, and 0.1 ml of 10% sodium nitroprosside solution, was added to 5 ml supernatant after centrifugation at 5000 rpm for 15 min. The tube was thoroughly shaken and the mixture was allowed to stand for 10 min. 25 ml of 3% aqueous solution of glycerine was added to the reaction mixture with frequent shaking over a period of 10 min. After additional 10 min interval, 2 ml of concentrated orthophosphoric acid was added drop wise to the mixture and the test tube was properly shaken. Colour development was allowed to produce for 5 min and colour intensity was measured at 540 nm in spectrophotometer (Perkin Elmer Lambda 68 UV VIS). The L-methionine yield was extrapolated from a standard L-methionine curve [15].
Recovery of L-methionine from fermented broth
An inexpensive down-stream recovery process that is capable of achieving the requisite recovery yield and purity is essential for producing any metabolite. Various levels of down-stream processing are required for the existing amino acid fermentation. The general approach to designing an efficient recovery scheme for bio products has been elucidated by Chisti and Moo-Young. The production scheme must accommodate the various regulatory requirements and consider the end use application of the product. Purification of L-amino acids relies on their physico-chemical properties, particularly solubility and isoelectric point. As the first step of the down-stream recovery process, the cells are separated from the fermentation broth by either centrifugation or filtration. The cell-free broth is then passed through activated charcoal columns for decolorization. L-methionine (isoelectric pH 5.74) can be recovered from the clarified broth by adjusting the pH to 5 with sulfuric acid to convert the amino acid to its cationic form and passing the broth though a bed of Amberlite IR-120 (H+) ion exchange resin at a controlled flow rate. The process is repeated until all the L-methionine is adsorbed. Afterwards, the column is washed with deionized water and eluted with 1(M) NH4OH to recover the L-methionine. Crystalline L-methionine can be obtained by concentrating under vacuum, treating with absolute alcohol, and drying overnight at 80°C [16-18].
Estimation of Dry Cell Weight (DCW)
The cell paste was obtained from the fermentation broth by centrifugation and dried at 1000C until constant cell weight was obtained [17-19].
Estimation of residual sugar: Residual sugar was determined by the DNS method as proposed by Miller [14].
Statistical analysis: All data were expressed as mean±SEM, where n=6, where ‘n’ denotes the number of experimental set up. Data were analyzed by one way ANOVA using a software Prism 4.0, considering p<0.05 as significant and p<0.01 as highly significant.
Response surface methodology: It consists of a group of experimental technologies, used for evaluation of relationship between different variables and measured responses. Plackett-Burmann design was used to assess the pick variables that influence L-methionine fermentation by the mutant significantly and insignificant factors were eliminated in order to obtain a smaller manageable set of variables. RSM was applied in two stages, first to trace out the significant variables for the production using Plackett-Burmann design criterion and later significant variables related from Plackett-Burmann design were optimized by a central composite design. The experimental design and statistical analysis of the data were done by using a software, prism 4.0.
Plackett-Burmann Design (PBD): Each variable was examined at two levels, namely a high level (+1) and low level (-1). Initial pH, volume of medium, age of inoculum, shaker’s speed, temperature, cell density, period of incubation, carbon source, nitrogen source, K2HPO4, KH2PO4, CaCO3, MgSO4.7H2O, NaCl, KCl, ZnSO4.7H2O, Na2MoO4.2H2O, MnSO4.4H2O, FeSO4.7H2O, biotin and thiamine-HCl were screened by conducting six experiments using Plackett-Burmann design. All experiments were conducted in six sets and mean values of L-methionine production was used for statistical analysis. The variables, which were significant at 1% level (p<0.01) from one way ANOVA were considered to have high impact on L-methionine production and were further optimized using Central Composite Design.
Central Composite Design (CCD): It was applied to determine the optimum levels of seven significant production parameters determined from PBD. The effects of the parameters (namely: age of inoculums, shaker’s speed, temperature, cell density, glucose concentration, nitrogen concentration, K2HPO4, KH2PO4, CaCO3, MgSO4.7H2O, FeSO4.7H2O and biotin) on L-methionine production by the mutant were examined at levels: -2, -1, 0, +1 and +2 where α+ 2n/4, where ‘α+’ represents the number of levels of significance considered (i.e., 5). Hence ‘n’ was the number of variables and ‘0’ corresponded to the central point. The level of each variable was determined by the following equation [20]:
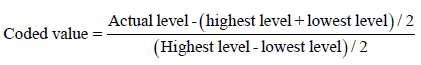 (1)
(1)
The experimental plan and the following independent variable were obtained from CCD: volume of medium, (15-35 ml), initial pH (6-8), age of inoculum (24-84), cell density (1 × 108-6 × 108 cells), temperature (25-32°C), period of incubation (24-108 h), carbon sources (glucose, fructose, sucrose, lactose, maltose, ribose, xylose, starch, dextrin, sodium citraite, sodium acetate and glycerol), different concentrations of glucose (20-140 g/L), nitrogen sources (urea, ammonium sulphate, sodium nitrate, ammonium chloride, ammonium nitrate, diammonium hydrogen phosphate, ammonium dihydrogen phosphate, ammonium carbonate, ammonium oxalate and ammonium citrate) and different concentrations of nitrogen (1-10 g/L) in the form of ammonium sulphate
After identification of significant variables using Plackett-Burmann design, Box-Wilson 24 factorial CCD was applied to optimize these variables. Five levels of variables were coded as [6]:
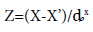 (2)
(2)
[Where, Z=Code value; X=Natural value; X’=Natural value in central domain; ȡx=increment of X corresponding to one unit of Z]. A total number of 31 experiments with 8 axial points (α=2) and six replications was applied for Box-Wilson 24 factorial CCD. The general form of the second degree polynomial equation was applied in this present study. The equation can be presented as follows [7]:
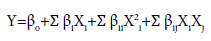 (3)
(3)
Where, Y=Response variable; βo=Coefficient of interaction effect (offset term); βi=Linear coefficient (ith term); βii=Coefficient of quadratic effect (iith term); βij=Interaction coefficient (ijth term). Analysis of variance (ANOVA) and regression analysis was done using software Prism 4.0.
Screening of variables by Plackett-Burmann design criterion for L-methionine production by Corynebacterium glutamicum X300
The variables which significantly affect the production of L-methionine by Corynebacterium glutamicum X300 were determined by Plackett-Burmann design. All the twenty one parameters such as volume of medium, initial pH, age of inoculums, shaker’s speed, temperature, cell density, period of incubation, glucose concentration, nitrogen concentration (in term of ammonium sulphate), KH2PO4, KH2PO4, CaCO3, MgSO4.7H2O, NaCl, KCl, FeSO4.7H2O, ZnSO4.7H2O, Na2MoO4.2H2O, MnSO4.4H2O, biotin and thiamine-HCl were examined at two widely spaced levels (Table 1).
| Code | Parameter | High level (+1) | Low level(-1) | ||
|---|---|---|---|---|---|
| Level | Activity (mg/ml) | Level | Activity(mg/ml) | ||
| I | Volume of medium (ml) | 35 | 8.8 ± 0.691 | 15 | 7.8 ± 0.881 |
| II | Initial pH | 8 | 7.9 ± 0.591 | 6 | 8.3 ± 0.779 |
| III | Shaker’s speed(rpm) | 300 | 8.6 ± 0.881 | 100 | 10.8 ± 0.728 |
| IV | Age of inoculum (h) | 84 | 4.8 ± 0.689 | 24 | 9.6 ± 0.913 |
| V | Cell density (cells) | 6×108 | 11.1 ± 0.881 | 1×108 | 6.3 ± 0.812 |
| VI | Temperature (°C) | 32 | 9.1 ± 0.681 | 25 | 4.8 ± 0.791 |
| VII | Period of incubation(h) | 108 | 8.6 ± 0.883 | 24 | 1.8 ± 0.681 |
| VIII | Glucose concentration (g/L) | 104 | 17.6 ± 0.992 | 20 | 9.6 ± 0.681 |
| IX | Nitrogen content (g/L) | 10 | 22.9 ± 1.181 | 1 | 16.1 ± 0.613 |
| X | K2HPO4(g/L) | 2.4 | 26.8 ± 1.631 | 1 | 23.9 ± 1.668 |
| XI | KH2PO4(g/L) | 2.1 | 33.8 ± 0.691 | 0 | 25.6 ± 0.916 |
| XII | CaCO3(g/L) | 3 | 37.3 ± 0.619 | 0 | 34.3 ± 0.793 |
| XIII | MgSO4.7H2O(g/L) | 1.8 | 28.1 ± 1.613 | 0.3 | 25.8 ± 0.874 |
| XIV | NaCl (g/L) | 2.4 | 42.8 ± 0.689 | 1 | 42.8 ± 0.613 |
| XV | KCl(g/L) | 2.4 | 40.8 ± 0.871 | 1 | 42.8 ± 0.932 |
| XVI | FeSO4.7H2O (mg/L) | 35 | 29.9 ± 0.993 | 5 | 26.6 ± 1.913 |
| XVII | ZnSO4.7H2O (mg/L) | 1.7 | 43.1 ± 0.793 | 0 | 40.2 ± 0.661 |
| XVIII | Na2MoO4.2H2O (mg/L) | 8 | 45.8 ± 0.881 | 0 | 44.2 ± 0.692 |
| XIX | MnSO4.4H2O (mg/L) | 6 | 46.7 ± 0.882 | 0 | 46.9 ± 0.692 |
| XX | Biotin (μg/ml) | 100 | 50.1 ± 0.832 | 0 | 48.1 ± 0.661 |
| XXI | Thiamine-HCl (μg/ml) | 100 | 51.2 ± 0.661 | 0 | 50.8 ± 0.913 |
Table 1: Level of variables examined for L-methionine production by the mutant Corynebacterium glutamicum X300 using Plackett-Burmann design criterion.
Among the twenty-one variables examined, shaker’s speed (III), age of inoculum (IV), cell density (V), temperature (VI), glucose concentration (VIII), nitrogen concentration (IX), KH2PO4 (X), KH2PO4 (XI), CaCO3 (XII), MgSO4.7H2O(XIII), KCl (XV), FeSO4.7H2O (XVI), MgSO4.7H2O (XIX) and biotin (XX) had significant (p<0.01) effect on L-methionine fermentation by the mutant, which was obtained from one way ANOVA. The coefficient of determinant (R2) of the model obtained from regression analysis was 0.861, suggesting thereby the model could explain up to 86.1% variation of the data. Thus the production of L-methionine by Corynebacterium glutamicum X300 using Placket_Burman design showed wide range of variations which implied that it required further optimization.
Application of Box-Wilson central composite design for the optimization of L-methionine production by Corynebacterium glutamicum X300
The optimum level of the key variables and the effect of their interactions on L-methionine production by the mutant were further examined using Central Composite Design (CCD) of RSM.
The first CCD
The fermentation trials for L-methionine by Corynebacterium glutamicum X300 were examined by CCD using the following variables: shaker’s speed (III), age of inoculum (IV), cell density (V), temperature (VI), glucose concentration (VIII), nitrogen concentration (IX), K2HPO4 (X), KH2PO4 (XI), CaCO3 (XII), MgSO4.7H2O (XIII), KCl (XV), FeSO4.7H2O (XVI), MnSO4.4H2O (XIX) and biotin (XX). The highest level of L-methionine was obtained up to 52.1 mg/ml with a biomass of 28.5 mg/ml and residual sugar content of 23.8%. Table 2 depicted the results of the second order Response Surface Models for L-methionine production by the mutant, obtained by one way ANOVA.
| Source | Degree of freedom (df) | Sum of square | Mean square | F-value | Probe˃F |
|---|---|---|---|---|---|
| Model | 82.169 | 7 | 9.313 | 18.313 | 0.0016 |
| Residual | 7.321 | 5 | 0.861 | 16.328 | 0.0616 |
| Lack of fit | 70.613 | 2 | 0.331 | 18.792 | 0.0516 |
| Pure error | 0.168 | 3 | 0.062 | 21.613 | 0.0476 |
| Total | 94.316 | ||||
R2=0.9982
Table 2: One way ANOVA for full quadratic model used for L-methionine production by Corynebacterium glutamicumX300.
Chi square test with a very low probability value (α<0.001) indicated that the model was highly significant. R2 (0.982) indicated that the sample variation for L-methionine production of 98.2% was attributed to the independent variables and only 1.8% of total variation cannot be justified by the model (Tables 3-5).
| Trial | Factor | L-methionine (mg/ml) [mean ± SEM] |
Predictedvalue (mg/ml) |
Residual values (mg/ml) |
|||
|---|---|---|---|---|---|---|---|
| X3 | X4 | X5 | X6 | ||||
| 1 | 150 | 48 | 6×108 | 31 | 13.6 ± 0.613 | 9.3 | 4.3 |
| 2 | 100 | 24 | 4×108 | 28 | 11.6 ± 0.998 | 10.1 | 1.5 |
| 3 | 100 | 72 | 4×108 | 25 | 10.8 ± 0.791 | 7.6 | 3.2 |
| 4 | 150 | 60 | 3×108 | 32 | 11.9 ± 0.991 | 9.1 | 2.8 |
| 5 | 100 | 36 | 5×108 | 28 | 13.2 ± 0.668 | 8.6 | 4.6 |
| 6 | 200 | 84 | 3×108 | 28 | 11.6 ± 0.792 | 9.3 | 2.3 |
| 7 | 150 | 60 | 6×108 | 27 | 13.3 ± 0.837 | 7.3 | 6.0 |
| 8 | 200 | 48 | 6×108 | 26 | 12.4 ± 0.991 | 8.1 | 4.3 |
| 9 | 100 | 48 | 3×108 | 30 | 14.8 ± 0.681 | 8.6 | 6.2 |
| 10 | 100 | 24 | 6×108 | 29 | 15.0 ± 0.927 | 9.4 | 5.6 |
| 11 | 150 | 72 | 6×108 | 32 | 14.2 ± 0.883 | 8.1 | 6.1 |
| 12 | 300 | 60 | 5×108 | 31 | 13.1 ± 0.813 | 7.6 | 5.5 |
| 13 | 150 | 84 | 4×108 | 31 | 13.6 ± 0.698 | 9.4 | 4.2 |
| 14 | 100 | 72 | 4×108 | 25 | 11.6 ± 0.735 | 7.8 | 3.8 |
| 15 | 300 | 72 | 1×108 | 29 | 11.3 ± 0.919 | 6.8 | 4.5 |
| 16 | 300 | 84 | 3×108 | 60 | 12.2 ± 0.882 | 9.6 | 2.6 |
| 17 | 200 | 84 | 3×108 | 60 | 13.6 ± 0.881 | 10.9 | 2.7 |
| 18 | 200 | 72 | 4×108 | 48 | 12.1 ± 0.802 | 9.3 | 2.8 |
| 19 | 300 | 48 | 5×108 | 96 | 10.8 ± 0.682 | 9.3 | 1.5 |
| 20 | 150 | 60 | 5×108 | 96 | 11.8 ± 0.792 | 7.6 | 4.2 |
| 21 | 100 | 48 | 1×108 | 26 | 11.3 ± 0.993 | 9.3 | 2.0 |
| 22 | 300 | 48 | 1×108 | 30 | 13.7 ± 0.813 | 10.6 | 3.1 |
| 23 | 150 | 60 | 5×108 | 30 | 12.6 ± 0.779 | 10.1 | 2.5 |
| 24 | 300 | 60 | 6×108 | 28 | 13.1 ± 0.681 | 6.1 | 7.0 |
| 25 | 200 | 72 | 3×108 | 28 | 14.3 ± 0.983 | 8.3 | 6.0 |
| 26 | 200 | 84 | 4×108 | 25 | 12.4 ± 0.779 | 9.6 | 2.8 |
| 27 | 200 | 84 | 4×108 | 26 | 11.7 ± 0.681 | 9.1 | 2.6 |
| 28 | 300 | 48 | 5×108 | 25 | 13.2 ± 0.832 | 10.4 | 2.8 |
| 29 | 200 | 48 | 3×108 | 26 | 11.4 ± 0.913 | 9.6 | 1.8 |
| 30 | 300 | 60 | 5×108 | 26 | 12.6 ± 0.872 | 8.3 | 4.3 |
| 31 | 200 | 72 | 1×108 | 30 | 11.3 ± 0.881 | 8.3 | 3.0 |
| 32 | 250 | 48 | 28 | 11.8 ± 0.692 | 7.9 | 3.9 | |
Table 3: L-methionine production by Corynebacterium glutamicumX300 using significant physical parameters based on CCD.
| Trial | Factor | Lmethionine (mg/ml) | Predicted value (mg/ml) | Residual Value (mg/ml) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIII | IX | X | XI | XII | XIII | XV | XVI | XIX | XX | ||||
| 1 | 40 | 2.0 | 1.2 | 2.0 | 3.0 | 1.2 | 0.5 | 20 | 0.0 | 00 | 49.6±0.913 | 46.8 | 2.8 |
| 2 | 20 | 10 | 2.3 | 1.9 | 2.0 | 1.8 | 1.0 | 5.0 | 1.6 | 80 | 41.6±0.772 | 37.6 | 4.0 |
| 3 | 140 | 7.0 | 1.4 | 0.0 | 1.5 | 0.6 | 1.0 | 20 | 6.0 | 2.0 | 36.3±0.832 | 31.2 | 5.1 |
| 4 | 40 | 7.0 | 1.0 | 1.2 | 2.5 | 0.3 | 0.0 | 35 | 2.5 | 1.6 | 50.1±0.661 | 42.8 | 7.3 |
| 5 | 60 | 1.4 | 1.8 | 1.0 | 2.5 | 1.8 | 2.0 | 20 | 2.0 | 4.0 | 52.2±0.992 | 44.6 | 7.4 |
| 6 | 40 | 1.0 | 2.0 | 2.1 | 3.0 | 0.3 | 2.0 | 35 | 3.0 | 4.6 | 50.3±0.683 | 46.1 | 4.2 |
| 7 | 60 | 1.4 | 2.2 | 1.8 | 3.0 | 0.6 | 1.0 | 5.0 | 3.0 | 2.6 | 49.6±0.813 | 44.3 | 5.3 |
| 8 | 120 | 2.0 | 2.1 | 1.6 | 1.5 | 0.3 | 1.0 | 5.0 | 4.5 | 4.1 | 48.2±0.669 | 41.6 | 6.6 |
| 9 | 80 | 1.4 | 2.1 | 1.6 | 1.0 | 1.5 | 1.6 | 10 | 2.0 | 6.1 | 50.3±0.832 | 46.4 | 3.9 |
| 10 | 40 | 2.0 | 2.0 | 2.0 | 2.0 | 1.2 | 1.7 | 20 | 4.0 | 4.8 | 46.8± | 41.6 | 5.2 |
| 11 | 120 | 1.0 | 1.6 | 1.6 | 1.6 | 1.0 | 1.0 | 20 | 0.0 | 3.9 | 31.3±0.661 | 28.3 | 3.0 |
| 12 | 140 | 4.0 | 1.8 | 1.8 | 0.0 | 1.5 | 1.4 | 10 | 2.0 | 3.3 | 42.8±0.779 | 39.3 | 3.5 |
| 13 | 40 | 1.0 | 1.8 | 1.6 | 2.0 | 1.8 | 0.0 | 35 | 2.5 | 0.0 | 36.8±0.681 | 32.2 | 4.6 |
| 14 | 40 | 1.0 | 1.0 | 0.0 | 1.5 | 0.6 | 0.5 | 25 | 2.0 | 0.0 | 41.6±0.832 | 38.9 | 2.7 |
| 15 | 60 | 4.0 | 2.0 | 0.0 | 2.0 | 0.9 | 0.0 | 25 | 4.0 | 20 | 46.6±0.881 | 41.3 | 5.3 |
| 16 | 80 | 1.0 | 2.3 | 1.6 | 1.5 | 0.9 | 0.0 | 10 | 6.0 | 5.3 | 48.9±0.732 | 44.2 | 4.7 |
| 17 | 120 | 1.0 | 2.1 | 1.6 | 1.5 | 0.9 | 0.0 | 10 | 6.0 | 3.9 | 41.3±0.913 | 38.3 | 3.0 |
| 18 | 60 | 6.0 | 2.3 | 1.8 | 1.5 | 0.3 | 2.0 | 10 | 4.0 | 4.6 | 46.2±0.883 | 42.8 | 3.4 |
| 19 | 100 | 7.0 | 1.0 | 2.1 | 3.0 | 0.6 | 2.0 | 10 | 4.0 | 4.3 | 46.3±0.961 | 43.1 | 3.2 |
| 20 | 140 | 1.0 | 1.0 | 2.1 | 3.0 | 1.8 | 1.5 | 5.0 | 2.5 | 4.1 | 44.8±0.832 | 41.6 | 3.2 |
| 21 | 100 | 1.4 | 2.3 | 0.0 | 3.0 | 1.8 | 1.5 | 5.0 | 4.0 | 4.3 | 50.1±0.599 | 46.2 | 3.9 |
| 22 | 120 | 9.0 | 1.6 | 0.0 | 2.5 | 1.8 | 1.7 | 5.0 | 2.5 | 3.9 | 40.8±0.599 | 38.3 | 2.5 |
| 23 | 120 | 1.0 | 1.4 | 0.0 | 0.0 | 1.2 | 1.0 | 20 | 4.0 | 3.1 | 46.2±0.832 | 41.6 | 4.6 |
| 24 | 140 | 8.0 | 1.0 | 1.0 | 1.0 | 1.8 | 1.5 | 25 | 6.0 | 4.8 | 44.6±0.611 | 41.1 | 4.6 |
| 25 | 60 | 1.0 | 1.0 | 1.6 | 1.5 | 1.2 | 1.5 | 20 | 6.0 | 4.1 | 50.1±0.432 | 46.2 | 3.9 |
| 26 | 60 | 1.4 | 1.6 | 1.2 | 2.0 | 1.5 | 1.0 | 20 | 2.5 | 3.6 | 39.7±0.568 | 37.1 | 2.6 |
| 27 | 80 | 1.0 | 2.3 | 1.6 | 2.0 | 1.6 | 1.8 | 35 | 2.0 | 20 | 48.3±0.662 | 44.2 | 4.1 |
| 28 | 60 | 9.0 | 1.8 | 1.4 | 2.5 | 1.4 | 1.5 | 1.6 | 6.0 | 30 | 41.6±0.881 | 39.3 | 2.3 |
| 29 | 100 | 6.0 | 2.3 | 1.8 | 1.0 | 1.8 | 1.5 | 1.4 | 5.0 | 20 | 43.3±0.793 | 40.1 | 3.2 |
| 30 | 120 | 2.0 | 2.2 | 1.2 | 0.0 | 1.2 | 1.2 | 1.6 | 6.0 | 10 | 44.6±0.801 | 39.6 | 5.0 |
| 31 | 140 | 1.4 | 2.3 | 1.6 | 1.5 | 1.6 | 1.8 | 0.0 | 4.5 | 10 | 48.6±0.872 | 44.2 | 4.4 |
| 32 | 60 | 9.0 | 2.1 | 1.6 | 1.5 | 1.6 | 1.5 | 2.4 | 6.0 | 20 | 40.1±0.661 | 38.1 | 2.0 |
Table 4: L-methionine production by Corynebacterium glutamicum X300 using significant nutritional parameters based on CCD.
| Variable | Coefficient | Standard error of mean | Computed t-value | p-value |
|---|---|---|---|---|
| Intercept | 52.168 | 0.039 | 233.611 | 0.000 |
| III | 0.913 | 0.061 | 1.169 | 0.016 |
| IV | 0.682 | 0.083 | 7.311 | 0.024 |
| V | 0.366 | 0.072 | 5.316 | 0.136 |
| VI | 0.611 | 0.091 | 2.618 | 0.613 |
| VIII | 0.732 | 0.059 | 4.813 | 0.024 |
| IX | 0.682 | 0.061 | 6.161 | 0.126 |
| X | 0.399 | 0.033 | 7.133 | 0.311 |
| XI | 0.816 | 0.042 | 4.613 | 0.791 |
| XII | 0.913 | 0.061 | 19.613 | 0.066 |
| XIII | 0.331 | 0.055 | 11.611 | 0.168 |
| XIV | -0.068 | 0.061 | -5.613 | 0.002 |
| XV | 0.611 | 0.066 | 6.918 | 0.069 |
| XVI | 0.382 | 0.079 | 9.832 | 0.361 |
| XIX | 0.791 | 0.069 | 8.622 | 0.162 |
| XX | 0.668 | 0.063 | 6.913 | 0.113 |
Table 5: Model coefficient calculated from linear regression for the assessment of the significance of the independent variables.
All total 32 experimental trials for both physical and nutritional variables were examined for the estimation of the principal effects and multi-factors interactions. The production of L-methionine was increased significantly (p<0.01) after optimization of nutritional parameters compared to the optimization of physical parameters.
The second CCD
Table 6 depicted the second order CCD for L-methionine production in the form of variance (ANOVA) for the quadratic model.
| Source of data | Sum of square | Degree of freedom (df) | Mean square | F-value | p-value˃F |
|---|---|---|---|---|---|
| Model | 0.841 | 5 | 0.313 | 1230.14 | <0.0001 |
| III | 0.797 | 6 | 0.331 | 226.64 | 0.0046 |
| IV | 0.401 | 3 | 0.210 | 86.46 | <0.0001 |
| V | 0.616 | 6 | 0.339 | 117.64 | <0.0001 |
| VI | 0.731 | 5 | 0.361 | 237.81 | 0.0024 |
| VIII | 0.611 | 3 | 0.305 | 373.61 | <0.0001 |
| IX | 0.383 | 1 | 0.116 | 423.64 | 0.0069 |
| X | 0.770 | 1 | 0.216 | 874.84 | 0.0011 |
| XI | 0.463 | 3 | 0.169 | 321.61 | 0.0022 |
| XII | 0.511 | 9 | 0.312 | 72.21 | <0.0001 |
| XIII | 0.663 | 3 | 0.226 | 123.81 | 0.0013 |
| XV | 0.634 | 3 | 0.213 | 321.21 | 0.0014 |
| XVI | 0.481 | 5 | 0.161 | 723.61 | <0.0001 |
| XIX | 0.514 | 1 | 0.226 | 54.33 | <0.0001 |
| XX | 0.681 | 4 | 0.261 | 222.21 | 0.0017 |
| Residual | 1.613 | 4 | 0.246 | - | - |
| Pure error | 0.000 | 4 | 0.000 | - | - |
| Lack of fit | 1.613 | 1 | 0.661 | - | - |
| Total | 10.66 | ||||
Table 6: One way ANOVA for Response Surface Quadratic Model for L-methionine production by Corynebacterium glutamicum X300.
Shih and Shen applied RSM to examine the yeast extract, glucose, ammonium sulphate and initial pH on the production of poly €-lysine by Streptomyces albulus IFO 14147 in shake-flask fermentation. They obtained both the first order with interactions model (R2=0.9660) which was more adequate than the pure first order model (R2=0.8759). They have applied CCD for the assessment of the optimum composition. Their experimental data were fitted with a second-order polynomial euation by a multiple regression analysis. The determination of coefficient (R2=0.816) and the Fisher’s test (significant at upper 5%) indicated a good adequacy of the secondorder polynomial model used to analyze the data [5]. Nilofer et al. used RSM to optimize the variables of L-lysine fermentation by Corynebacterium glutamicum AEC-2. They used Plackett-Burman design and obtained four variables (the level of sugar in molasses, ammonium sulphate, and incubation temperature and inoculum size) were proved to be significant for L-lysine production. Furthermore, CCD (24 factorial) was applied to determine the optimum levels of significant variables. A second order polynomial regression model was used to explain the experimental data. Using these models, the production of L-lysine was increased up to 2.6 fold [6]. Pandey and Banik, optimized six factors (namely: pH, temperature, fermentation time, orbital speed, age of inoculum and inoculums volume) for alkaline phosphate production by Bacillus licheniformis using RSM. They have reported that pH, temperature, fermentation time and orbital speed were significant (p<0.05) using Placket Berman design methodology. An increase up to 1.5 fold in the production was obtained after optimization of the production using RSM [7]. Shankar et al. examined the invertase production by Saccharomyces cerevisiae MK. The optimum levels of the key variables (orange peel, yeast extract and methionine) was applied to determine their interactions on the production using CCD and RSM. The determined coefficient of determination (R2=0.9994) was nearer to 1 which satisfied the adjustment of the quadratic model to the experimental data [1]. Patel et al. compared between one at a time variation factors and CCD for the production of mycophenolic acid by Penicillum brevicompactum MTCC8010 in a 12-day batch culture. The medium optimization using one-at-a-time variation gave 6 fold greater titer, whereas CCD gave almost 9 fold greater titer compare to the production prior to the optimization [21].
The nearness of the coefficient of determinant (R2=0.9982) for one way ANOVA for full quadratic model and the same for one way ANOVA for Response Surface Quadratic Model (R2=0.9991) used for L-methionine production by Corynebacterium glutamicum X300 ensured the satisfactory adjustment of the Quadratic model to explain the data obtained from the present experiment. The maximum production of L-methionine (52.1 mg/ml) was obtained with 72 h of incubation.