Journal of Proteomics & Bioinformatics
Open Access
ISSN: 0974-276X
ISSN: 0974-276X
Research Article - (2015) Volume 0, Issue 0
Background: This study aimed to find the linker length with minimal impact among chains to fight against the structure and function of cervical cancer single-chain antibody.
Methods: The original variable region of heavy chain (VH) and variable region of light chain (VL), and the singlechain antibody with (Gly4Ser)n linkers of different lengths (n=1~8) were modeled using SWISS-MODEL homologous modeling. Comparison of the similarity of original VH/VL and VHn/VLn was carried out by applying the algorithm of spatial hierarchical alignment based on the spherical coordinates. The fore and aft distance and diffusion radius of α were also calculated. The stability of antibody with different linker length was then compared. ELISA method was adopted to evaluate the immunological activity of single-chain antibody with different linkers. MTT assay was used to analyze the inhibition effect of ScFv-n on cervical cancer cells.
Results: When linker unit n=4, the structures were the most similar between ScFv and the original VH/VL. When n=3, the similarity of it had little differences to that when n=4, and the influence of adding connecting peptide on the stability of single-chain antibody was the least, when n=3. The result of ELISA and MTT methods indicated that when n=3, single-chain antibody gained the highest activity.
Conclusion: The optimum length of linker of anti-human cervical cancer single-chain antibody was n=3 from the point of mathematical modeling and biology experiments. This study provided new ideas for the design and constructions of single-chain antibody, and theoretical basis for the treatment of cervical cancer.
Keywords: Cervical cancer; Single-chain antibody; Linker; Homologous modeling; Similarity algorithm
Cervical cancer (CC) is a malignant tumor occurred in the uterus vagina and cervical canals. It is one of the most common gynecological malignancies [1] and the incidence takes the second place among gynecologic oncology. Each year, there are about 530,000 new cervical cancer cases worldwide, of which 85% occur in developing countries, and cervical cancer is one of the main causes of death of women in developing countries [2-4]. Therefore, how to improve the long-term survival of patients with cervical cancer is an important issue in clinical and basic research.
With constantly thorough study of molecular biology, genetically engineered antibody has been widely used in the diagnoses and treatments of diseases. It has great potential in the diagnosis and treatment of malignancy. Single-chain variable fragment (ScFv) is a small molecule antibody that has intact antigen-binding site. Compared with the defects of monoclonal antibody, such as large amount of antibody molecules, low penetrating ability, low clearance rate of blood and low ration of tumor blood [5], ScFv has advantages, such as small amount of antibody molecules, high penetrating ability, low immunogenicity and easy to mass production. It can also neutralize virus and toxin to give play to immunization in cells and be guided carrier. However, studies about the application of ScFv in CC treatment are rarely reported. As a result, it is of particular significance to conduct an in-depth research on the single-chain antibody of CC, which would provide a new direction of treating CC.
ScFv is recombinant protein linked by a length of connecting peptide through VH and VL. Different lengths of linkers among chains have different influence on the active of single-chain antibody, so an insight into the impact of different lengths of linkers among chains on the biological activity of single-chain antibody has great significance in the establishment of single-chain antibody and screening of linkers among chains. By using the bioinformatics method, this study compared the impact of different linkers among chains on the structures and functions of single-chain antibody of CC from the aspects of protein tertiary structure, aiming to providing new ideas of the screening of interchain linker length of CC and the establishment of single-chain antibody, and also providing some theoretical bases for the targeted therapy of CC.
Experimental materials
The amino acid sequences of single-chain antibody VH and VL of CC were from the articles related to researches of the anti-human cervical cancer written by Wang et al. [6]. Hela human cervical epithelial cell line was preserved in our lab. We ourselves constructed the general expression vector of single-chain antibody of connecting peptide with different length, expressed by HB2151 expressing system, and ammonium sulfate was used for preliminary purification.
Experimental methods
Selection of linker: By adding a suitable linker between VH and VL, the amino acid sequence of single-chain antibody could be obtained. At present, the most widely used linker, proposed by Huston et al. [7], was (Gly4Ser)n with a rigid structure. That is, 5 amino acid residues consisted of n-times occurred four glycine and a serine [8,9]. The peptide chain structure had high flexibility (i.e., the ability of spatial bending), and small spatial steric hindrance, which was in favor of the interaction between VL, VH fragment as well as formation of the correct conformation of them, and improving the stability of antibody molecules. The structure would not be recognized and degraded easily by protease, and was conducive to the internal stability of the antibody molecule.
Modeling: (Gly4Ser)n (n=1, 2, 3…) was adopted as the linker of the single-chain antibody in this study. ScFv-n was modeled using SWISSMODEL homologous modeling (http://beta.swissmodel.expasy.org/interactive) and the structured data file and three dimensional (3D) structured diagrammatic figures of corresponding VHn and VLn were obtained. At the same time, we modeled a three-level structure of VH and VL. Thus the pdb structured data file (pdb code: VH:2gki; VL:3hzm) and 3D structured diagrammatic figures corresponding to original VH and VL could be obtained, which were treated as the reference data of structural similarity comparison.
Similarity analysis: This study consulted the space spherical shell hierarchical matching algorithm based on spherical coordinates, which is put forward by Zhang et al. [10] to calculate the similarity. First, coordinate of 3D structure model that provided by pdb file was converted into spherical polar coordinates using formula (1, 2, 3).
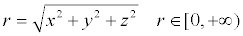 (1)
(1)
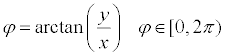 (2)
(2)
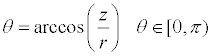 (3)
(3)
Then the largest space of molecules was separated into spherical shell unit with certain radial thickness. The number of the same kind of atoms in every spatial block was calculated to build vector quantity a, b. The amount of the same kind of atoms (mainly C, N, and O) in different space shell was counted and summed up in accordance with predetermined weight. The last number of atoms in that shell was obtained. Multi-dimensional vector ai and bi was constituted by the number of atoms in every shell according to the hierarchical division order. At last, the commonly used similarity included angle cosine of approximate function was conducted as the approximate function of similarity. It was showed as following:
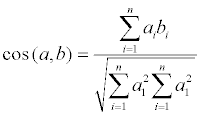
Stability analysis of spatial structure: The stability of ScFv could be expressed by the fore to aft distances or the diffusion radiuses, which means the stabilities of single-chain antibody with different lengths of linkers were compared by the distances from first alpha carbon atom to the last alpha carbon atom and diffusion radiuses. Matlab was adopted to establish the positions of original VH, VL, and alpha carbon atom path structure of ScFv in the space coordinates system. The fore to aft distances of VH and VL original structure and that of VHn and VLn when added different length of linker (n=1~8) were figured out. Then their maximum diffusion radiuses were calculated, respectively.
Gene identification of single-chain antibody: Single-chain antibody expression vectors constructed by connecting peptide containing different length were identified using PCR. PCR products were analyzed using 1.0% agarose gel electrophoresis.
ELISA detection of single-chain antibody immunological activity: Human Hela cells in the logarithmic phase were collected. They were inoculated in 96-well plates of 5 × 104 cell/mL, 100 μL/well. It was cultured at 37°C for 24 h, washed 3 times with PBS, and fixed by 2.5% glutaraldehyde at 37°C for 5 min. After washing, 100 μL (0.50 mg/mL) single-chain antibody of connecting peptide with different length was added to each well as the primary antibody. PBS was added in blank control well. Goat-anti-Mouse HRP antibody was treated as the second antibody. 100 μL antibody was added to each well, and incubated at 37°C for 2 h. Substrate TMB was added after washed with PBS, coloration at 37°C away from light for 5 min. Then 100 μL stop buffer (1 mol/L H2SO4) was added to each well. Microplate reader was used to detect the OD value at a wavelength under 450nm.
Detection on single-chain antibody inhibition rates for cervical cancer cells using MTT assay: Human Hela cells in the logarithmic phase were selected, whose concentration were then transferred to (2 ~ 4)×105 cells/mL, inoculated in 96-well plates of 100 μL/well, and cultured in 5% CO2 at 37°C. Each well in the experimental group was added 100 μL (0.50 mg/mL) single-chain antibody containing a different length of the connecting peptide. Each length of the Linker was made in triplicate. One group of cancer cell suspension without antibody was set up as control group, and a group added culture medium was regarded as zero hole, cultured in 5% CO2 cell incubator at 37°C. Each well was added 100 μL MTT (0.2 mg / mL), incubated for 4 h at 37°C, and removed the supernatant. 200 μL of it was then added to alkalize DMSO (10% glycine -NaOH buffer), incubated for 1 h, vibrated, and determined the A value at 570 nm using microplate reader. Each well was repeated more than three times, and the results were basically the same, with the median test values as the criterion. The inhibition rate of cervical cancer cells represented the activity levels of a single-chain antibody. The formula is as follows:
Inhibition rate=(1-A value in experimental group/A value in control group)×100%
Original VH and VL three dimensional structures of ScFv
The original VH and VL were modeled by SWISS-MODEL. The result was shown as Figure 1.
Three dimensional structures of ScFv-n (n=1~8)
In general, the linkers of ScFv were ranged within 4 to 44 amino acids [11]. The longer the linker was, the more instable structure of ScFv was. In the process of modeling, when linker unit n=9 (the linker was 45 amino acids in length), the searched template had lower homology, so the modeling declared a failure. Therefore, we finally obtained the pdb data file and 3D model of ScFv-n when n=1~8, which meant the lengths of linkers were ranged within 5-40 amino acids. The 3D structures were presented in Figure 2.
Figures 1 and 2 had shown that the original structural model of VH and VL had some differences to the structure after adding the linkers. In addition, the structure of the linker was changed following the length of it. It was visible that the lengths of these added linkers had direct influence on the spatial structure of single-chain antibody.
Similarity analysis of VH and VL of ScFv-n
The similarities among the original VH/VL and VHn/VLn with adding linkers were compared using protein similarity algorithm based on spherical polar coordinates hierarchy. The similarities were showed in Table 1.
| Number of layers | 2 | 3 | 4 | Mean value | Number of layers | 2 | 3 | 4 | Mean value | Total mean value |
|---|---|---|---|---|---|---|---|---|---|---|
| VH1 | 0.9203 | 0.8419 | 0.6324 | 0.79820 | VL1 | 0.9943 | 0.9709 | 0.8867 | 0.9506 | 0.8744 |
| VH2 | 0.9143 | 0.8419 | 0.6042 | 0.78680 | VL2 | 0.9932 | 0.9709 | 0.8953 | 0.9531 | 0.8700 |
| VH3 | 0.915 | 0.8412 | 0.6175 | 0.79123 | VL3 | 0.9932 | 0.9709 | 0.8953 | 0.9531 | 0.8722 |
| VH4 | 0.9117 | 0.8423 | 0.6463 | 0.80010 | VL4 | 0.9932 | 0.9709 | 0.8953 | 0.9531 | 0.8766 |
| VH5 | 0.9171 | 0.8416 | 0.6365 | 0.79840 | VL5 | 0.9932 | 0.9697 | 0.8849 | 0.9493 | 0.8738 |
| VH6 | 0.9154 | 0.8419 | 0.6157 | 0.79100 | VL6 | 0.9932 | 0.9709 | 0.8953 | 0.9531 | 0.8721 |
| VH7 | 0.9142 | 0.8386 | 0.6474 | 0.80007 | VL7 | 0.9938 | 0.9697 | 0.8942 | 0.9526 | 0.8763 |
| VH8 | 0.9151 | 0.8391 | 0.6521 | 0.80210 | VL8 | 0.9932 | 0.9697 | 0.886 | 0.9496 | 0.8759 |
Table 1: Comparative results of similarity.
During the counting process, it was indicated that the sequence of single-chain antibody was fairly short. If the number of molecules’ shells was too large, the results would not be stable and correct enough. In this way, spherical molecules were separated into 2, 3 and 4 layers. Then the similarities were calculated and the mean value was obtained. The higher similarity indicated the minimal impact of adding linkers on the structure of protein, and the minimal impact on the activity of single-chain antibody. Table 1 showed that when linker unit n=4, the similarity between that VH/VL and original VH/VL was the highest, which indicated that the influence of the adding linkers on the structures of single-chain antibody in variable region was the least.
Stability analysis of ScFv-n
The structure of the path of alpha carbon atom of original VH, VL and ScFv-n that built by Matlab 2008 was compared with positions in Figures 3 and 4. The paths of alpha carbon atom present the fore and aft distance changes of VHn and VLn more directly, indicating that changes occurred in alpha carbon atom path structure, but small changes in overall radial of molecules after adding linker with different lengths.
The influences on the stability of single-chain antibody after adding linkers were analyzed through the calculation of the fore and aft distances and diffusion radiuses. The results were shown as Table 2.
| Distance between head and tail | Radius | Distance between head and tail | Radius | Distance between head and tail | Radius | Distance between head and tail of linkers | |||
|---|---|---|---|---|---|---|---|---|---|
| VH1 | 53.6011 | 34.7554 | VL1 | 46.7338 | 30.0575 | VHL1 | 45.6794 | 216.5253 | 10.1888 |
| VH2 | 56.2322 | 38.7563 | VL2 | 43.6335 | 27.6651 | VHL2 | 45.6783 | 216.5592 | 14.0992 |
| VH3 | 55.6829 | 39.5128 | VL3 | 43.6429 | 27.6653 | VHL3 | 45.6783 | 216.8067 | 11.7021 |
| VH4 | 50.1406 | 37.4041 | VL4 | 43.6302 | 27.6654 | VHL4 | 45.6783 | 215.9425 | 27.8454 |
| VH5 | 45.9529 | 28.5646 | VL5 | 43.6084 | 27.6612 | VHL5 | 45.6782 | 216.3861 | 14.2882 |
| VH6 | 56.1668 | 37.7859 | VL6 | 43.6321 | 27.6654 | VHL6 | 45.6777 | 217.4843 | 12.2691 |
| VH7 | 48.304 | 31.8512 | VL7 | 43.6263 | 27.6624 | VHL7 | 45.6782 | 217.0619 | 12.7475 |
| VH8 | 45.406 | 30.8712 | VL8 | 43.6034 | 27.6596 | VHL8 | 45.6803 | 216.3476 | 26.4597 |
| VH | 39.9954 | 41.8673 | VL | 43.9975 | 29.1371 |
Table 2: Distance between head and tail and radius of VH and VL before and after adding Linkers.
The results in Table 2 showed that:
i. When linker unit n=8, the fore and aft distance of VH was the closest to that of original VH. While n=3, the diffusion radius of VL was the closest to that of original VL. When n=1, the diffusion radius of VL was the closest to that of original VL. Moreover, when n=2~8, the diffusion radiuses of VL were almost the same. As a result, when n=3, the influence of adding linker on the stability of single-chain antibody was the least.
ii. The fore and aft distances and radiuses of the whole protein molecule changed little after adding linkers.
iii. When n ≤ 4, the fore and aft distances of linkers were increased with the increase of n value. However, when n>4, the fore and aft distances of linkers were decreased for the spatial folding and bending.
Gene identification of single-chain antibody
The established expression vectors of single-chain antibodies with different lengths of Linkers (n=1~8) were identified. After analyzing PCR products by agarose gel electrophoresis, an apparent DNA fragment at about 830 ~ 950bp was obtained (Figure 5). The sequencing results were entirely consistent with the expected sequence.
ELISA detection of single-chain antibody immunological activity
ELISA method was used for quantitative test the immunological activity of single-chain antibody and results showed that, when linker unit n=1 ~ 8, all single-chain antibody had specific binding activity to cervical cancer cells. When n<3, specific binding activity was low; when n=3, the activity was the highest; when n>3, the activity decreased in different degrees. The results were shown in Figure 6.
MTT method detection of inhibition rate on cancer cells of single-chain antibody
MTT method was used to detect the inhibiting effect of single chain antibody with different of Linkers on HeLa cell line. The results were shown in Figure 7. When linker unit n=1 ~ 8, the inhibition rate was more than 20%. When n=3, the inhibition rate of ScFv against cervical cancer cell was the highest, reaching 68%. When n>3, the inhibition rate decreased slightly.
Single-chain antibody (ScFv) is a new reorganized protein, which belongs to a micro molecule antibody of genetic engineering antibody. It has characters like small number of molecules (around 27 kD, only 1/6 of the complete antibody), no Fc fragment, low immunogenicity, good affinity, strong penetrability, short half-life period in body circulation, and easy to reform by genetic engineering, etc. It has an extensive application prospect, because it has become the oriented carrier of diagnoses and treatments of tumors. Zitzmann et al. [11] took advantages of phage antibody library technology to screen the ScFv (DUP21) of prostate specificity, and the imaging showed that the radioactivity of cancerous area of prostate was three times higher than the normal tissue when marked by I131, which indicated that ScFv possessed favorable targeting. DeNardo reported that the combined application of ScFv that marked by IFN-C and I could increase part of the mark concentration of tumors up to 2-4 times, which can remarkably strengthen the imaging results [12].
(Gly4Ser)n link peptide in this study, proposed by Huston [7], can not only link the N-terminal to C-terminal in V region, but can tighten the VH and VL without affecting the interaction between them. However, the linker length of heavy variable region (VH) and light variable region (VL) will affect the stability and activity of the single-chain antibody. Too short linker may affect the freedom of spatial folding of protein, thus affecting the structure and function of protein; while the over-long linkers will affect the stability of ScFv, increase the immunogenicity and produce anti- heterologous protein response [13-15]. So the selection of linker is very important. The linker should possess limpness, not form steric hindrance to interference the folding of heavy chains, and not have bad influence on the solubility of products. Most of the applications of single-chain antibody targeted drug were still at stage of in vitro and animal experiments, while they had many difficulties in application to human body [16], of which the most important was that the affinity of ScFv was lower than the parent McAb. As to this problem, we can change the link peptide among chains to increase the affinity of ScFv.
Methods in inferring the biomolecules functional similarity mainly include two categories, one is protein comparison based on sequence; the other is structural and functional analysis based on structure. The bioinformatics believes that molecular structure determines the property and function of the molecule, and that the spatial structure of the protein determines its biological function. Therefore, by comparing the spatial structure of protein, this study aimed to compare the similarity of protein molecules [17]. First, the changes of the structures of variable regions as the lengths of linkers change were analyzed using theoretical 3D modeling. The study found that when linker unit n value was 4, the similarity of between ScFv and original structure was the highest. The similarity of n=3 was little different from that of n=4. By calculation of the distances from the first alpha carbon atom to the last alpha carbon atom and diffusion radiuses, it was found that the influence of linker on the stability of single-chain antibody in the variable region was the least when n value was 3. Through indirect ELISA and MTT methods, it was found that the biological activity of single-chain antibody was much higher when n value was 3.
So far, there have been many studies about the influence of linker length on the activity of ScFv. Gustavsson [18] proposed that linkers to construct single-chain bispecific antibodies was 4 to 44 amino acids in length. Le et al. [19] proposed that the range of the length of link peptides was within 6-27 amino acids. Weisser et al. [20] found that when the number of amino acids of linkers was 15, that is, n=3 in (Gly4Ser)n, the affinities, activities and the other aspects could reach a satisfactory level. All these were in an agreement with the results of the present study.
To sum up, the results of this study indicated that when n value of linker was 3, the structure and biological activity of anti CC singlechain antibody were ideal, and (Gly4Ser)3 could be regarded as the linker of single chain antibody against human cervical cancer, which provided a basis for further study on anti-human cervical cancer singlechain antibody. It was found that the three-dimensional structure of the protein is more stable than its primary structure. The present study compared the stability of the protein in terms of protein structure. Through the three-dimensional structure model of the protein and the use of spherical polar coordinates, we calculated the similarity of the protein and analyzed the stability of the protein structure, thus this study provided a new idea on protein function analysis as well as the design and construction of single-chain antibody, and laid the foundation for the targeted therapy of cervical cancer.
We thank to the Medical Engineering Technology and Date Mining Institute of Zhengzhou University. This institute has given us a great deal of help in experimental guidance and advice. Then, we would like to thank the First Affiliated Hospital of Zhengzhou University. This hospital offered us a perfect experimental base and date sources. Thank you!