Advanced Techniques in Biology & Medicine
Open Access
ISSN: 2379-1764
ISSN: 2379-1764
Research Article - (2017) Volume 5, Issue 4
Statement of the problem: When molecular biologists observed in 1953 that the sequence of the DNA four bases in the nucleus of a cell influenced the sequence of the twenty amino acids of protein in the cytoplasm, they desired to find a code to account for the correlation, and eventually had the 64 triplet genetic code in 1954 from a mathematician, which is currently in use but not flawless.
Methodology and theoretical orientation: The said observation is seen as a natural example of an input/output system, in which the input is the DNA four bases and the output is the sequence of 24 permutations of the four DNA bases constituting the genetic code in the cytoplasm. A combinatorial input/output multiplicative replication system armed with basic permutations computation schemes is now available to produce permutations systematically, such as Square Kinematics Scheme and Successive Collateral Posting Scheme used.
Findings: A 24 quadruplet genetic code was produced by each of the two methods with an input set of the DNA four bases. It is shown in the successive collateral posting method that the 64 triplets comprise 40 non-permutations and 24 permutations. The 40 non-permutation triplets are crossed out leaving 24 permutation triplets which are undersized and therefore unqualified to represent the genetic code output sequence from an input set of 4.
Conclusion and significance: The 24 quadruplet genetic code is a breakthrough in the Molecular Biologists’ search for a code following their observation which ended up with the 64-triplet genetic code that has no combinatorially valid code word being triplets, instead of quadruplets.
Recommendations: These are made towards effective publicity of the new 24 quadruplet genetic code to attract experimental experts to spell it to win adoption in coding application in protein studies for a desired relief to stakeholders in genetics.
Keywords: 64-triplet genetic code; Molecular biology; Cytoplasm; Protein synthesis studies
The 64-triplet genetic code structure that begged for revisit is immature and a mixture of 24 permutation and 40 non-permutation triplet codons derived in 1954 by the base-4 neo-digibreed indirect method with input set of RNA four bases A, U, G, C (Adenine, Uracil, Guanine, Cytosine). The revisit is from combinatorial perspective, because the task of deriving a genetic code structure from the given four nucleotide bases is in the province of computational combinatorics and borders on the generation of permutations of 4 from 4 (Figure 1).
The objectives of this work are four-fold, given the challenge to derive afresh a code from the four RNA bases, A, U, G, C (Adenine, Uracil, Guanine, Cytosine) to account for the correlation between them and twenty amino acids of protein as sequel to molecular biologists’ observation in the early 1950s, that the sequence of the four nucleotide bases A, U, G, C in the nucleus of a cell influenced the sequence of the 20 amino acids of protein in the surrounding cytoplasm in the cell, because the one of 64-triplet structure based on 43 derived in 1954 in response to the scientists’ quest and spelt thereafter and adopted since 1968 in coding application in protein synthesis studies is bedeviled with irregularities which are only widely discussed in genetics literature, but without remedy to date, hence the first objective is to derive a genetic code without any irregularity using the same material and indirect method used in 1954 by the authors of the current one now in disfavour. The second objective is to use a direct method to produce the irregularity-free genetic code for confirmation of validity. The third one is to show why the 64-triplet genetic code structure is bedeviled with the well-known irregularities. Lastly, the fourth objective is to highlight the combinatorial and molecular biological merits of the new genetic code structure of 24 quadruplets and thereby attract spelling experts to spell it in order to render it fit for adoption for coding application in protein synthesis studies, in the event of the publication of this work as per this attempt, and that to the relief of all stakeholders worldwide.
Materials
The materials consist of the RNA four bases in the sequence of A, U, G, C (Adenine, Uracil, Guanine, Cytosine) as transcription from the DNA four bases in the order of A, T, G, C (Adenine, Thymine, Guanine, Cytosine) as carried in a particular rung of the double helix and are used as input set of 4 bases in the multiplicative replication input/output system in computational combinatorics developed by this author in the 1990’s.
Methods
The methods are two, namely: base-four neo-digibreed indirect method, Chart 1 and Square kinematics direct method, Chart 2 under Figure 2.
Chart 1: Derivation of 24 quadruplet genetic code structures from the four RNA bases A, U, G, C by successive collateral posting method using base four neodigibreed and de-isodigitation.
Note: The 24 quadruplet digitisms standing in lines 22 to 85 represent the valid genetic code structure in agreement with combinatorics Summary of valid permutation codons
Calculated value4P4=4! =4 × 3 × 2 × 1=24 quadruplets
Computed value 4P4=24 quadruplets
First, base-four neo-digibreed indirect method uses an input set of the RNA four bases in the linear sequence of A,U,G,C involving successive collateral posting (SCP) in Fati’s Geotropic Frame to the limit of 44 where 256 quadruplets are produced. This product is a mixture of 24 permutation quadruplets (crops) bearing no repetition(s) of letters, and 232 non-permutation quadruplets (weeds) bearing repeated letters as carried in Chart 1 in lines 22-85. The 232 non-permutation quadruplets are crossed out as weeds to leave a residue of 24 permutation quadruplets as crops for harvesting as the valid codons.
Direct method, designated as square kinematics: The input set of RNA four bases A, U, G, C are loaded at the corners of the square in clockwise direction as depicted in Figure 3. The loaded square is deployed in three ways as depicted in Figures 3a-3c, to generate 8 combinatorially valid quadruplets per deployment per section using kinematics and view mixing as shown in Chart 3 carrying a genetic code structure of 24 quadruplet codons from lines (1) to (24).
Chart 3: Protein type proliferation and diversification, climax of merits of quadruplets codons.
Note: The 24 quadruplet digitisms standing in lines 22 to 85 represent the valid genetic code structure in agreement with combinatorics Summary of valid permutation codons
Calculated value 4P4=4! =4 × 3 × 2 × 1=24 quadruplets
Computed value 4P4=24 quadruplets
A genetic code structure of 24 permutation quadruplets is presented in Table 1 as a computational reality for the result, being a combinatorial derivation with the two sources or methods stated. The list of 20 amino acids of protein is adapted from the book [1].
| Input set of RNA four bases | Output permutations of quadruplet codons | Remarks | |||||
|---|---|---|---|---|---|---|---|
| Serial no. of 24 quadruplet codons | 20 amino acidsa/4 signals | Methods | Some salient points | ||||
| Amino acids 20/signals 4 to be specified by codons upon spelling by the experimental experts | (2)Indirect | (2) Direct | |||||
| Base-four neo-digibreed, Ref. Chart 2 lines 22-85 | Square Kinematics, Ref. Chart 3 lines 1-24 | ||||||
| [1] | Genetic Code Sequence | Genetic Code Sequence | |||||
| AUGC | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 |
Alanine Arginine Asparagine Aspartic acid Cysteine Glutamic acid Glutamine Glysine Histedine Isoleucine Leucine Lysine Methionine Phenylalanine Proline Serine Threonine Tryptophan Tyrosine Valine Signal 1 Signal 2 Signal 3 Signal 4 |
YTBD “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ “ |
AUGC AUCG AGUC AGCU ACUG ACGU UAGC UACG UGAC UGCA UCAG UCGA GAUC GACU GUAC GUCA GCAU GCUA CAUG CAGU CUAG CUGA CGAU CGUA |
AUGC CGUA UGCA ACGU UAGC UACG CAUG GUAC AGUC UCGA UCAG GACU GAUC CAUG CUGA AGUC AUCG GCUA UAGC CGAU ACUG GUCA CAGU UGAC |
(1) Output sequence of permutations per method is unique. (2) All 24 codons per sequence possess integrity and potency. (3) Collinearity between 24 codons and 20 amino acids/4 signals evident. (4) All codons are convertible to equivalents of DNA rungs in base content by the replacement of U by T showing that the genetic code is actually the RNA transcribed from the DNA as intimated [2]. |
|
| Total | 24 | 24 | 24 | 24 | 24 | ||
Table 1: New genetic code of 24-quadruplet codon structure.
(aList of 20 amino acids of protein adapted from Figure. 17.4, The World of Cell, p.529 by Becker, Wayne M. (1986). b By Jill Wright et al (1988) in their book, Prentice Hall Life Science at page 63 with regard to protein synthesis, where it is stated that the RNA in the ribosomes, along with the RNA sent out from the nucleus directs the production of proteins)
YTBD: Yet To Be Determined; Signal 1: Place Start Signal; Signal 2: Time Start Signal; Signal 3: Place Stop Signal; Signal 4: Time Stop Signal
This discussion is geared to exploring the significance of the results of the work in the context of the four objectives of the revisit set out in the introduction as follows:-
i. Derivation of a genetic code without any irregularity, using the same material and indirect method used in 1954 by the authors of the current 64-triplet code;
ii. Using the same material of RNA four bases A, U, G, C to produce an irregularity-free genetic code structure by a direct method for confirmation of validity;
iii. To show why the 64-triplet genetic code is bedeviled with the much publicized irregularities; and
iv. To highlight the combinatorial and molecular biological merits of the new genetic code structure of 24 quadruplet codons and thereby attract spelling experts to spell it in order to render it fit for adoption in coding application, and that, to the relief of all stakeholders worldwide.
The significance of the result of 24-quadruplet genetic code structure recorded in Figure 4 under item (i) above is, it is concise and precise. The difference in quantum in terms of codewords and base units from 64 codewords to 24 code words, i.e., a reduction to less than half and in base units from 64 × 3 equal to 192 to 24 × 4 equal to 96, showing a reduction of base units involved to exactly half, is clear. The significance here boils down to less labour, besides combinatorial accuracy. Secondly, inspection of Chart 1 lines 6-20 will reveal some code words amongst columns A, U, G, C crossed out, which upon counting would be 40 in number. They are infested with isodigitation, i.e., repetition of digits (bases) and therefore belong to the category of digitism (code words at this instance) known in computational combinatorics as nonpermutations. The number of code words surviving the crossing out in the designated region of the chart upon counting would be 24. They are free from isodigitation or base repetition(s) and are said to belong to the category of code words known in combinatorics as permutations. Only permutations are qualified for engagement in the genetic code structure which is a natural example of input/output multiplicative replication system in computational combinatorics. So the distinction made between permutations (crops, bearing no repeated letters) and nonpermutations (weeds, bearing some repeated letters) amongst the 64 triplets produced by the indirect method of successive collateral posting (SCP) is a remarkable significance in the derivation of the true genetic code structure from the input set of A, U, G, C. The 24 permutation triplets surviving the crossing out or de-isodigitation, nevertheless, are immature and undersize relative to the input set of 4 and are therefore disqualified as code words, hence the chart is continued to the digitality level 4 combination covering lines 22-85 wherein 256 quadruplets are carried. Of these only 24 are permutations (crops) and are standing as residue for harvesting, while 232 are crossed out being non-permutations (weeds). The distinction between permutations and non-permutations has therefore helped to yield 24 valid code words from a list of 256 quadruplets produced by the indirect method of SCP or the base-4 neo-digibreed indirect method of permutation production (Chart 1). Therefore another significance of the result under item (i) is the offer of a reliable indirect method or technique for deriving the true genetic code structure of 24 quadruplets of the status of permutation. If we might consider the results of the work in terms of software and hardware, then under item (ii) above, the significance of the result of the work can be identified as the provision of additional direct method or combinatorial technique (hardware) for deriving a correct 24-quadruplet genetic code structure (software) from the same material input set of RNA four bases A, U, G, C with precision as per Chart 2 under Figure 2. Thus the square kinematics technique for direct production of the true genetic code structure is of much significance in molecular biology in the context of protein synthesis studies. With the 24-quadruplet structure confirmed by an independent method, the validity of the derivation result is assured, and it is definitely a breakthrough in the derivation of the combinatorially correct genetic code structure. Under item (iii) above: “To show why the 64-triplet genetic code is bedeviled with the much publicized irregularities” as per Figure 4, the significance of the result of the work (revisit) is the identification and elimination of the forty non-permutation triplets as combinatorial irregularities of the genetic code structure as the combinatorial output of the input set of RNA four bases A, U, G, C, by the indirect base-4 neo-digibreed method.
Combinatorial error is detected in the derivation of the 64-triplet genetic code structure from the four RNA bases A, U, G, C, as shown in Figure 5. The source of the error is traced to the wrongful interpretation of selections for permutation as neo-digibreed population formula (bd), where b is base-strength or input set and d is the digitality required, instead of permutation factorial complements for set (n) and selection (r) given by (r) from (n), i.e., 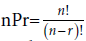 as depicted in Figure 4: “The parted ways of right and wrong at selections for permutation in the derivation of genetic code from input set of four RNA bases in box-graphics”.
as depicted in Figure 4: “The parted ways of right and wrong at selections for permutation in the derivation of genetic code from input set of four RNA bases in box-graphics”.
The significance of greater importance is that the new 24-quadruplet genetic code structure is a corrective measure freed of all the irregularities associated with the 64-triplet structure and doubling as a refutal to the degenerate code to the end that there is no room again for hypotheses of the kind of the Wobble [3] phenomenon. The derivative is now a true copy of the genetic code engaged in protein synthesis in plants and animals, flawless, since creation and upholding the inerrancy of Nature all the way.
Lastly, under item (iv) above, on highlighting the combinatorial and molecular biological merits of the new genetic code structure of 24 quadruplets of permutation status, the significance of the result of the work (revisit) in molecular biology as conveyed in Chart 3 “Protein type proliferation and diversification, climax of merits of quadruplet codons” is two-fold. Firstly, the material is easily adapted to demonstrate the working of the observation in the early 1950s by molecular biologists that, the sequence of the four RNA bases A, U, G, C, in the nucleus of a cell, influenced the sequence of the 20 amino acids of protein in the surrounding cytoplasm of the cell as per Chart 3 in which Column 1 bearing the input set A, U, G, C, at the top and the output of 24 quadruplets is meant to represent the nucleus of a cell, while Column 2 carrying the various output sequences of the input sets in numbered rows 1-24 is meant to represent the cytoplasm of the cell. There are two subrows per numbered horizontal chamber in Column 2. The upper subrow carries the output sequence of the genetic code structure corresponding to the input set in the adjoining chamber in Column 1. The lower subrow per numbered horizontal chamber in Column 2 carries the related protein type with four interspersed codon-sized empty spaces left by four signals 1-4 for time and place based start/stop controls in protein synthesis. The configuration of the chart ideally consists of odd rows to the right and even rows to the left of Column 1 in order to represent the neighbourhood of nucleus and cytoplasm in the cell, but it is altered to what is seen in the chart, because of space constraint.
Secondly, Chart 3 carrying 24 protein types (in the lower sub rows) is an illustration of ABC of protein type proliferation and diversification. Chart 3 is evidence of the efficiency of the new genetic code of 24-quadruplet structure which can boast of collinearity (Chart 3, Column 2). Two other molecular biological merits are that each quadruplet codon is identifiable with a specific amino acid or control signal and is capable of reproducing the entire genetic code structure when deployed as input set. Both functions are exercisable in furtherance of protein synthesis. Two more molecular biological merits of the new genetic code structure hinged on codon integrity are the workability of both Chargaff’s rules of A=T or U and G=C [4] and Watson-Crick’s base pairing of A/T or U and G/C [5] as illustrated in Diagram 1 captioned “New Genetic Code Structure in Dendritic Dichotomization”, where 2(A=C=12) lines and 2(G=C=12) lines per genetic code sequence of 24 quadruplet codons bearing 96 nucleotide base units in four kinds in keeping with Chargaff’s rules. Also base pairing of 2 (A/U × 12) lines and 2 (G/C × 12) lines per genetic code sequence of 24 quadruplets codons featuring 96 nucleotide base units in four kinds in conformity with Watson-Crick’s rules is apparent. Yet another remarkable molecular biological merit of the new genetic code structure is that it represents any portion of the DNA double helix comprising 24 consecutive rungs in base-content as depicted in Chart 3 Column 1, when U is replaced by T. The combinatorial merits are exemplified by codon integrity, codon potency, and codon deployability as input set in the input/out multiplicative replication system of computational combinatorics, all owed to permutation status as shown in Figure 6, titled, Combinatorial essentials of theoretical plant flow-chart for protein synthesis and primary folding of protein type in box-graphics. ”It is also noteworthy that the work has asked and answered a fundamental question in protein synthesis, “Why the genetic code in protein synthesis?” as per Figure 6 parts 1 and 2. Hence the genetic code option to justify its adoption in nature must be the workforce of 24 quadruplet codons that it is with collinearity and repeatability for continuity and sustainability that can match the unending task of protein synthesis in plants and animals since creation.
Figure 6a: One of two options for input set in box-graphics protein synthesis, why the genetic code? Protein synthesis: one of two options concerning deployment of 20 constituent amino acids of protein as input set for permutation for combinatorial alignment in the production of protein types; self or substitute:
Collinearity evident in the one-to-one correlation between the 24 quadruplet code words and the 20 amino acids of protein and 4 control signals in protein synthesis in plants and animals as portrayed in Chart 3 is the sure evidence of the combinatorial correctness or accuracy of this new genetic code combinatorial structure yielded by this revisit of the degenerate 64-triplet genetic code structure Figures 7a and 7b. This new genetic code version is wholeheartedly recommended to experimental experts in molecular biology etc. for spelling in order to render it fit for coding application in protein synthesis studies.
• Let it be noted at this point that the 64-triplet genetic code structure begging for combinatorial revisit is immature and a mixture of 40 non-permutation and 24 permutation triplets which are as incompatible as weeds and crops during harvest.
• The genetic code structure of 24 permutation quadruplets is the combinatorial answer to the raising of enough code words from the four-letter alphabet of the RNA four bases A, U, G, C. (Adenine, Uracil, Guanine and Cytosine).
• The diversification of protein types follows from the variation of sequence of 24 output factorial complements being the code words as influenced by changes of sequence of bases in the input set of four.
• With a quadruplet input set of RNA bases whether in the indirect method of base-4 neo-digibreed scheme or direct method of Square Kinematics, the combinatorially correct output factorial complements of 4 from 4 can only be 24 permutation quadruplets given by factorial 4=4!=4×3×2×1=24 quadruplets.
• The new genetic code structure of 24 quadruplets affords collinearity, one-to-one correspondence, with the 20 amino acids of protein/4 control signals for effective protein synthesis characterized by protein type diversification.