Research Article - (2018) Volume 4, Issue 2
The Effects of Pantoea and Kosakonia Isolated from Buckwheat Sprouts on Obese Mice.
*Corresponding Author: Kanako Yamanouchi, Department of Health Sciences, Hirosaki University, 66-1 Hon-cho, Hirosaki, 036-8564, Japan, Tel: +81-172-39-5973, Fax: +81-172-39-5973 Email:
Abstract
Objective: This study aims to investigate the effects of surface bacteria of buckwheat sprouts on obesity. We examine whether these bacteria have probiotic properties.
Methods: Since P-36 and P-37 strains were present in large amounts on the buckwheat sprouts surface, identification of species was carried out by intestinal bacteria identification kit and 16S rRNA gene analysis. Subsequently, to investigate the biological effects, P-36 and P-37 strains were divided into two groups, heat-treated dead bacteria and untreated viable bacteria, which were then orally administered to obese model mice exhibiting hyperglycemia every other day for a total of 10 times. After administration, BMI and visceral fat mass were measured, and oral glucose tolerance test was conducted to evaluate glucose tolerance; their influence on blood glucose level and insulin resistance (HOMA-IR) was evaluated.
Results: The strain P-36 was identified as Pantoea sp., and P-37 as Kosakonia cowanii. In the group of mice administered the living and dead bacteria of P-36 and P-37, the weight gain rate after the administration was low, and the progression of obesity was suppressed. In the oral glucose tolerance test, an improvement of glucose tolerance and a significant suppression of blood glucose level was confirmed at 0 and 15 min in the group treated with viable P-37. In addition, HOMA-IR was improved in mice treated with both viable and dead P-37 bacteria.
Conclusion: The mice administered P-36 and P-37 showed a gradual increase in body weight and a decrease in visceral fat percentage. The mechanism leading to improved blood glucose levels observed in mice administered P-37 strain remains unclear at present. The involvement of buckwheat sprouts-derived bacteria in improving hyperglycemia and reducing obesity during a short time will help to discover new probiotics.
Keywords: Buckwheat sprouts; Surface microbial flora; Pantoea; Kosakonia; Obesity; Hyperglycemia; Insulin resistance
Introduction
Buckwheat sprouts are a traditional vegetable in the Tsugaru Region of Aomori Prefecture, Japan, and they have been traditionally grown since over 350 years ago. Recent studies on rutin and anthocyanins present in buckwheat and their health benefits have been attracting increased attention to buckwheat sprouts as a new functional vegetable [1-3]. However, growing as well as shipping of many types of sprout vegetables cultivated in hydroponic cultures is challenging as frequently bacteria develop on their surfaces [4-6]. Therefore, we have sterilized the surface of buckwheat sprouts grown in hydroponic culture and subsequently analyzed the bacterial flora in an attempt to extend the expiration date for consumption and facilitate safe food distribution. By chance, it was found that the buckwheat sprout surface bacteria belonging to Enterobacteriaceae comprised a significant percentage of the characteristic bacterial flora, in particular the genera Pantoea.
Pantoea spp. are relatively newly delineated bacterial genera segregated from the genus Enterobacter [7]. About 20 species of Pantoea are presently known, some of which are used as biocontrol products for promotion of plant growth, suppression of fungi, and other [8,9]. In addition, Pantoea promote the activation of innate immunity, improve allergic dermatitis and activate intestinal immunity in rainbow trout, and their role in diabetes, anticancer action, and activation of immunity has been investigated [9-11].
Therefore, P-36 and P-37 strains were isolated from buckwheat sprouts and bacterial species were identified. In additon, to investigate the biological effects of P-36 and P-37 strain on metabolic processes in living organisms, we studied the influence of these two strain on obesity in mice.
Materials and Methods
Isolation of bacteria
Determining the number of viable bacteria in buckwheat sprout: In accordance with Food Hygiene Test Guidelines [12], 225 mL of physiological saline was added to 25 g of buckwheat sprouts, and the mass was kneaded for 1 min to make a 10-fold diluted stock solution. A 101- to 106-fold diluted solution was prepared from this diluted stock solution, and 20 μL of each diluted solution was smeared on a standard agar medium, followed by aerobic culture at 35°C for 48 h; viable bacteria colonies were counted for each sample. Colony count was repeated twice for each diluted solution.
Bacterial species identification: About 50 bacterial colonies that grew on the agar medium with smeared buckwheat sprout wash solutions were selected to identify bacterial species using a kit for identifying intestinal bacteria (API 20E; Sysmex Biomérieux Co., Ltd., Japan). Among them, the strains P-36 and P-37, both of which had a large number of colonies, were examined for morphological characteristics under an optical microscope, and catalase reaction, oxidase reaction, acid/gas production from glucose, and oxidation/fermentation of glucose was surveyed for each of the two strains (the first stage test) [13]. The property test (the second stage test) was carried out using a kit for intestinal bacteria identification API 20E (Sysmex Biomérieux). Based on the results of gene analysis and bacterial property analysis, additional tests, which included growing bacterial cultures at 4°C and 10°C, on a potassium cyanide medium, and in the presence of L-rhamnose, were conducted for species identification.
Gene analysis of isolated P-36 and P-37: DNA extraction of P-36 and P-37 strains were performed with achromopeptidase (Wako Pure Chemical Industries, Japan), and a PrimeSTAR HS DNA Polymerase kit (Takara Bio, Japan) was used for PCR gene amplification with primers 9F and 1510R [14]. Sequencing reactions were performed using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) and decoded in an Applied Biosystems 3130 xl Genetic Analyzer System (Applied Biosystems) using primers 9F, 785F, 536R, 802R, 1242R, and 1510R and ChromasPro 2.1 software (Technelysium Pty. Ltd., AUS) [14]. BLAST search of obtained sequences was performed in DB-BA 12.0 (TechnoSuruga Laboratory, Japan) and the international nucleotide sequence database (DDBJ/ENA (EMBL)/GenBank) [15]. Sequences were edited in BioEdit ver 7.2 and aligned in CLUSTAL W[16,17]. Molecular phylogenetic estimation using neighbor-joining method was performed using MEGA ver 7.0, and reliability of the tree topology was evaluated by bootstrap analysis with 1000 iterations [18-21].
Pantoea administration experiment
Creation of obesity model mouse: Five-week-old C57BL/6NJcl female mice were obtained from CLEA Japan, Inc. (Japan). The mice were acclimatized for 3 weeks after acquisition before the experiment. They were fed 10 kcal% fat diet as a control feed group and 60% fat meal (Research Diets, Inc., USA), and their weight and feed intake were measured over time. After 4 weeks of feeding, the mice were deprived of any food for 6 h, and after measuring their body weights, they were subjected to a 2 g/kg glucose tolerance test [22]. Because a significant increase in blood glucose level was confirmed in mice fed 60% fat diet as compared with the control feed group, they were used as an obesity model mouse for the experiment. The animals were kept in a room at a temperature of 23.9 ± 0.3°C and humidity of 26.6 ± 4.2%. The experiments with the animals were conducted in compliance with the guidelines concerning laboratory animals at Hirosaki University (Experimental animal approval number: G 14007).
Preparation of administered bacterial solution: P-36 and P-37 strains (Pantoea sp.) cultured overnight at 30°C on a heart infusion (HI) agar medium were transplanted into 100 mL of HI liquid medium and cultured with stirring at 30°C for 16 h. After culturing, the precipitate was recovered by centrifugation at 8,500 × g and 4°C for 30 min. The recovered sediment was washed twice with phosphate buffered saline (PBS (-), pH 7.2) suspended in 2 mL PBS to prepare the bacterial solution. Bacterial solution contained viable bacteria. To prepare a solution with dead bacteria, the bacteria were killed by boiling at 100°C for 10 min and then cultured for 48 h on a standard agar medium to confirm they were not viable.
Calculation of the number of bacteria to be administered: After preparing the bacterial solution, the number of bacteria was calculated each time it was administered to the mouse. To this end, the prepared bacterial solutions were diluted 107-1010, and 20 μL of each diluted solution was smeared on a standard agar medium, and aerobically cultured at 30°C for 24 h to count viable bacteria. Colony count was repeated twice for each diluted solution.
Administration of P-36 and P-37 bacterial fluids to mice: The mouse experimental groups were divided into a PBS-administered healthy mouse group (control group; n=6), PBS-administered control group fed 60 kcal% fat meal (Ob-PBS group; n=6), obese group administered viable P-36 (Ob-P-36 group; n=5), obese group administered dead P-36 (Ob-HTP-36 group; n=5), obese group administered viable P-37 (Ob-P-37 group: n=5), and obese group administered dead P-37 (Ob-HTP-37 group: n=5). Bacterial fluid was administered starting from the fifth week of 60% fat diet ingestion, when mice showed symptoms of hyperglycemia. Bacterial fluid was administered 10 times every other day, and a glucose tolerance test was conducted at 2 days after the final administration. The bacterial solution was administered at 2.1 × 1010 ± 5.0 × 109 CFU/200 μL/mouse for P-36, and at 2.4 × 1010 ± 3.8 × 109 CFU/200 μL/mouse for P-37.
Glucose tolerance test (oral glucose tolerance test), homeostatic model assessment for insulin resistance (HOMA-IR): Five grams of D-(+)-glucose (FUJIFILM Wako Pure Chemical Corporation, Japan) was suspended in physiological saline and the volume was adjusted to 25 mL by adding the saline. The solution was administered to mice that were fasted for 6 h using an oral probe in amounts equivalent to 2 g/kg [22]. Blood was collected by tail vein puncture and glucose level was measured using gluco card G black (ARKRAY, Inc., Japan) (double measurement). Glucose levels were measured 0 min before and 15 min, 30 min, 60 min, and 120 min after glucose administration. In addition, HOMA-IR was calculated using the formula: (fasting glucose level × fasting insulin level)/22.4 [22]. The insulin level was measured with an ultra-sensitive mouse insulin measurement kit (Morinaga, Japan) from blood serum.
Body mass index (BMI) measurement: After completion of administration, a glucose tolerance test was conducted and the body length was measured under isoflurane anesthesia. The length of the mouse was measured from the abdominal side, from the nose tip to the tail root. BMI was calculated as BMI=w/t2, where w is body weight (kg) and t is height (m).
Measurement of visceral fat: Euthanasia was performed by cervical dislocation on the third day after administration, and the weight of visceral fat was measured. Visceral fat was calculated as the sum of the weights of the mesenteric adipose tissue and perirenal adipose tissue, and the value was expressed as the visceral fat percentage of the body weight.
Statistical analysis: The differences in blood glucose levels between groups was assessed by analysis of variance and Tukey test; significant difference was assumed at P<0.05 Student's t test was used to compare the administration group and the obesity group (PBS) in each experiment. All statistical analyses were conducted in Origin Ver. 8.1 (OriginLab, USA).
Results
Number and type of bacteria living on buckwheat sprouts
The surface bacteria of the commercialized soya bean was 8.9 × 106 CFU/mL. The proportion of strains showing properties of Pantoea spp. according to API 20E analysis was 75.5%. Strains P-36 and P-37, which were present in highest density within those Pantoea spp., were selected and used for the experiment.
P-36 and P-37 were motile and gram-negative bacilli, able to ferment glucose, and had a positive catalase reaction, but negative oxidase reaction (Table 1). These properties are consistent with the properties of Enterobacteriaceae [23].
| Items | P-36 | P-37 | ||
|---|---|---|---|---|
| Culture temperature (°C) | 30 | 30 | ||
| Cell morphology | Bacillus (0.8–0.9 × 1.5–2.0 μm) |
Bacillus (0.8–0.9 × 1.5–2.1 μm) |
||
| Gram stainability | - | - | ||
| spore | - | - | ||
| Motility | + | + | ||
| Colony morphology | Culture: nutrient agar Culture time: 24 h Diameter: 1.0–2.0 mm Color tone: yellow Form: circle Elevated state: lenticular Colony edge: entire Surface: smooth Transparency: opaque Consistency: butter-like |
Culture: nutrient agar Culture time: 24 h Diameter: 2.0–3.0 mm Color tone: yellow Form: circle Elevated state: lenticular Colony edge: entire Surface: smooth Transparency: opaque Consistency: butter-like |
||
| Growth temperature(°C) | 37 | + | + | |
| 45 | - | + | ||
| Catalytic reaction | + | + | ||
| Oxidase reaction | - | - | ||
| Acid/gas production from glucose (acid production/gas production) | +/+ | +/+ | ||
| O/F test (oxidation/fermentation) |
+/+ | +/+ | ||
| +: Positive, -: Negative | ||||
Table 1: First step test of isolated bacterial strains.
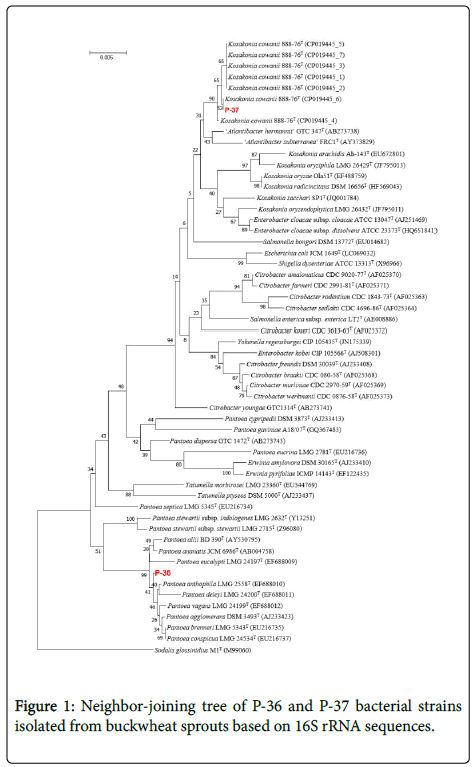
The results of the bacterial second stage test conducted using the API 20E kit revealed that P-36 was able to reduce nitrate and showed β-galactosidase activity, had no lysine decarboxylase, ornithine decarboxylase, and urease activity, produced indole and acetoin utilizing citric acid, did not hydrolyze gelatin, and could ferment D-mannitol, D-sorbitol, and amygdalin, but not inositol (Table 2). This strain developed colonies when cultured at 10°C and in the presence of L-rhamnose as a fertilizer (Table 3). These properties are similar to those of Pantoea anthophila, and phylogenetic analysis based on 16S rDNA sequences placed it within a clade with other Pantoea species (Figure 1), but do not properties of bacteria agree [24]. Therefore, we labeled this strain as Pantoea sp.
| Items | P-36 | P-37 |
|---|---|---|
| β-galactosidase * | + | + |
| Arginine dihydrolase * | - | - |
| Lysine decarboxylase * | - | - |
| Ornithine decarboxylase * | - | - |
| Usability of citric acid * | + | + |
| H2S production * | - | - |
| Urease * | - | - |
| Tryptophan deaminase * | - | - |
| Indole Production * | + | - |
| Acetoin production * | + | + |
| Gelatinase * | - | - |
| Glucose ** | + | + |
| D-Mannitol ** | + | + |
| Inositol ** | - | - |
| D-sorbitol ** | + | + |
| L-rhamnose ** | + | + |
| White sugar ** | + | + |
| D-Melibiose ** | + | + |
| D-amygdalin ** | + | + |
| L-arabinose ** | + | + |
| Oxidase * | - | - |
| NO2 production * | + | + |
| Reduction to N 2 gas * | - | - |
| Motility | + | + |
| Growth on MacConkey agar medium | + | + |
| Oxidation in OF medium * | + | + |
| Fermentation in OF medium * | + | + |
| * Biochemical test, ** Fermentation / oxidation test | ||
Table 2: Second stage test of isolated bacterial strains.
| Items | P-36 | P-37 |
|---|---|---|
| Growth at 4°C | - | |
| Growth at 10°C | - | |
| Growth in KCN | +w | + |
| Assimilability of L-rhamnose | + | |
| +: Positive, -: Negative, w: Growth is weak | ||
Table 3: Adding Tests based on the result of 16S rRNA region sequence and Second stage test of the isolated strain.
P-37 strain reduced nitrates, exhibited β-galactosidase activity, and showed no arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, and urease activity. In addition, it produced acetoin but not indole, did not hydrolyze gelatin, and was able to ferment D-mannitol and L-rhamnose, but not inositol (Table 2). This strain did not grow at 10°C, but cell cultures were developed on potassium cyanide medium (Table 3). These properties nearly corresponded to Kosakonia cowanii. This was further corroborated by the phylogenetic analysis, which resolved P-37 within the clade with other K. cowanii isolates (Figure 1) [25]. Therefore, we concluded that this strain belongs to K. cowanii.
Mouse weight change and visceral fat
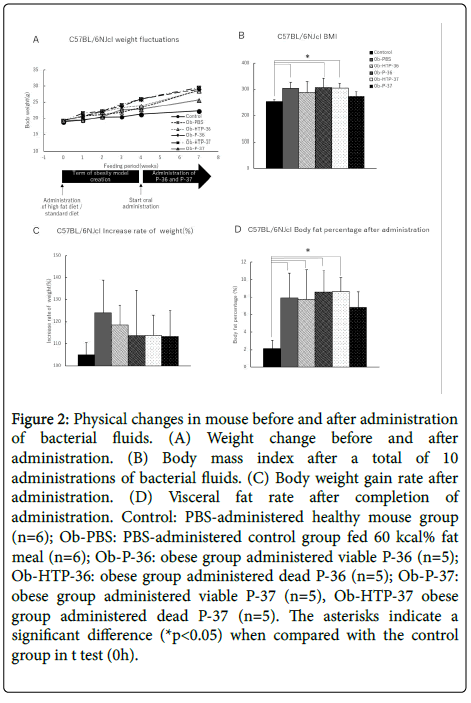
Body weight of the experimental mouse group did not decrease within 3 weeks from the start of administration of bacterial fluid (Figure 2A). Furthermore, After bacteria administration, BMI of all treatment groups increased compared with the healthy mouse group, except for the groups administered with viable P-37 strains, which did not exhibit any decrease in obesity levels (Figure 2B). However, when comparing weight gain rates before and after administration of bacterial fluids, the rate of weight gain showed a decreasing trend in all treated groups compared to the obese group (Figure 2C). In addition, the visceral fat percentage showed a slight decrease in mice administered live P-37 (Figure 2D).
Figure 2: Physical changes in mouse before and after administration of bacterial fluids. (A) Weight change before and after administration. (B) Body mass index after a total of 10 administrations of bacterial fluids. (C) Body weight gain rate after administration. (D) Visceral fat rate after completion of administration. Control: PBS-administered healthy mouse group (n=6); Ob-PBS: PBS-administered control group fed 60 kcal% fat meal (n=6); Ob-P-36: obese group administered viable P-36 (n=5); Ob-HTP-36: obese group administered dead P-36 (n=5); Ob-P-37: obese group administered viable P-37 (n=5), Ob-HTP-37 obese group administered dead P-37 (n=5). The asterisks indicate a significant difference (*p<0.05) when compared with the control group in t test (0h).
Blood glucose level fluctuation and HOMA-IR
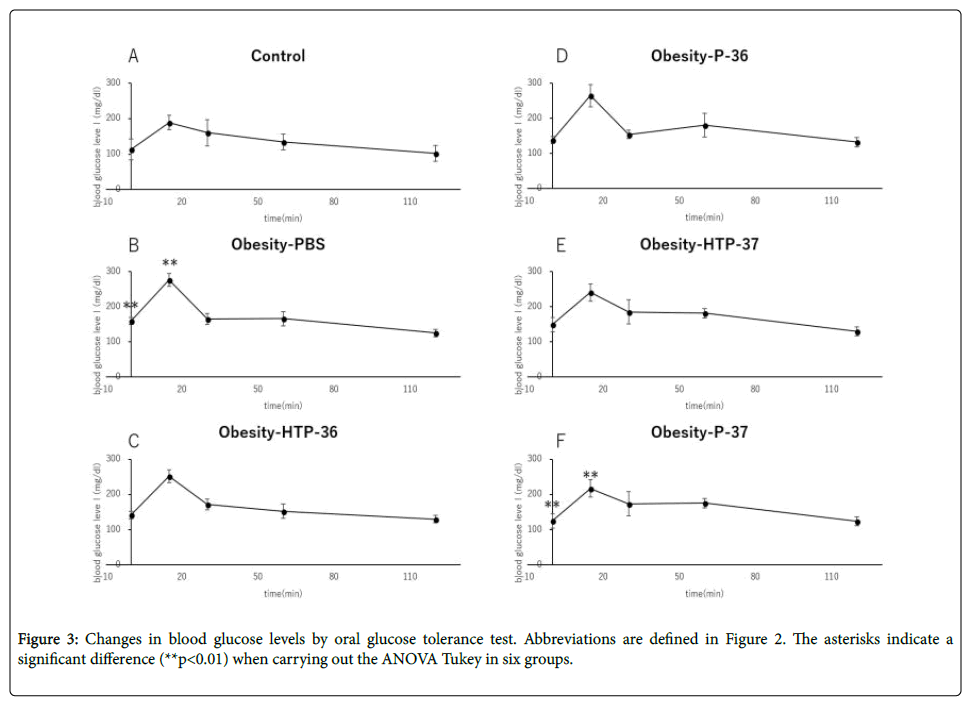
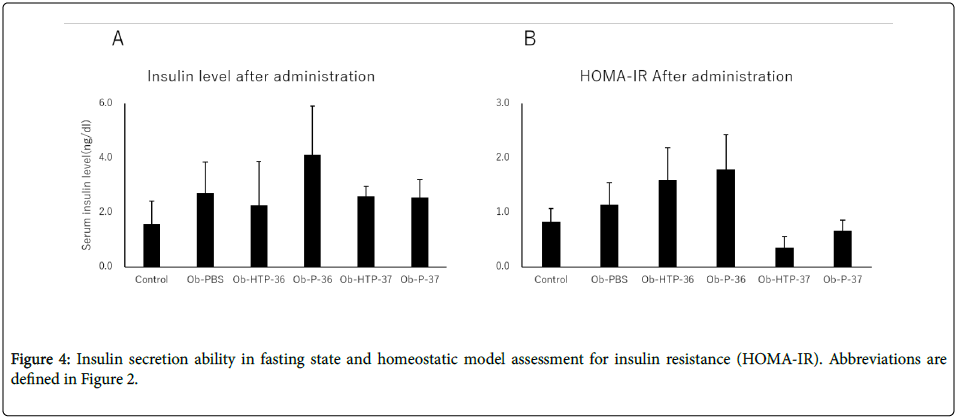
Compared to healthy mice (Control), Obesity-PBS group predominantly showed elevated blood glucose levels at 0 and 15 min after the start of glucose test. In the groups administered bacterial fluids, the blood glucose level suppression effect was confirmed, although the degree of suppression varied between the groups at 15 min from the test start (Figures 3A-3F). In the group administered viable P-37, blood glucose level was suppressed significantly compared with the obese group at 0 and 15 min of the glucose text (Figure 3F). There was no significant fluctuation in fasting insulin levels in both the obese group and the groups administered bacterial fluids (Figure 4A). However, HOMA-IR improved in the groups administered viable and dead P-37 (Figures 4A and 4B).
Discussion
In the present study, we identified two strains belonging to Enterobacteriaceae isolated from buckwheat sprouts and assessed their effect on obesity in mice. P-37 identified as Pantoea by analysis with the intestinal bacteria identification kit (API 20 E) was Kosakonia cawanii according to 16S rRNA analysis. Various microorganisms such as root nodule bacteria and fungi are engaged by plants, where they promote nutrient absorption of plants, impart dry stress tolerance, and improve resistance to pathogens [26,27]. Pantoea and Kosakonia identified herein are also known to coexist with plants and facilitate plant growth through nitrogen fixation [9,11]. However, reports on their biological effects are scarce. The present study is the first to report about improvement of glucose tolerance and HOMA-IR by these strains. Improved insulin resistance is associated with improvement of type 2 diabetes caused by obesity [28], and the change in insulin resistance is known to have a strong interaction with the intestinal flora [29]. Although the mechanisms underlying improved glucose tolerance and HOMA-IR need to be further elucidated and the optimal dose of bacteria is yet to be determined, we can conclude that the involvement of Pantoea and Kosakonia derived from buckwheat sprouts are a potentially new probiotic discovery.
Hyperglycemia and hyperlipidemia due to obesity, inflammatory reaction of the whole body, and other conditions have been recently associated to a change in intestinal flora [30-32]. Therefore, it is necessary to investigate how oral ingestion of P-36 and P-37 affects the intestinal flora and to elucidate the mechanism of their action.
Financial Supports
This work was supported by the Aomori Genki corporate challenge grant project 22 (Research fund number 109 ) and Hirosaki University GOGO fund (Research fund number 109).
References
- Kim SJ, Maeda T, Sarker MZ, Takigawa S, Matsuura-Endo C, et al. (2007) Identification of anthocyanins in the sprouts of buckwheat. J Agric Food Chem 55: 6314-6318.
- Kimi SL, Son YK, Hwangi JJ, Kim SK, Hur HS, et al. (2001) Development and utilization of buckwheat sprouts as functional vegetables. Fagopyrum 18: 49-54.
- Tsurunaga Y, Takahashi T, Katsube T, Kudo A, Kuramitsu O, et al. (2013) Effects of UV-B irradiation on the levels of anthocyanin, rutin and radical scavenging activity of buckwheat sprouts. Food Chem 141: 552-556.
- MRobertson LJ, Johannessen GS, Gjerde BK, Loncarevic S (2002) Microbiological analysis of seed sprouts in Norway. Int J Food Microbiol 75: 119-126.
- Dechet AM, Herman KM, Parker CC, Taormina P, Johanson J, et al. (2014) Outbreaks caused by sprouts, United States, 1998-2010: lessons learned and solutions needed. Foodborne Pathog Dis 11: 635-644.
- Kim SA, Kim OM, Rhee MS (2013) Changes in microbial contamination levels and prevalence of foodborne pathogens in alfalfa (Medicago sativa) and rapeseed (Brassica napus) during sprout production in manufacturing plants. Lett Appl Microbiol 56: 30-36.
- Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P (2013) Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36: 309-319.
- Feng Y, Shen D, Song W (2006) Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J Appl Microbiol 100: 938-945.
- Walterson AM, Stavrinides J (2015) Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev 39: 968-984.
- Skalli A, Castillo M, Andree KB, Tort L, Furones D, et al. (2013) The LPS derived from the cell walls of the Gram-negative bacteria Pantoea agglomerans stimulates growth and immune status of rainbow trout (Oncorhynchus mykiss) juveniles. Aquaculture 416-417: 272-279.
- Berger B, Baldermann S, Ruppel S (2017) The plant growth-promoting bacterium Kosakonia radicincitans improves fruit yield and quality of Solanum lycopersicum. J Sci Food Agric 97: 4865-4871.
- Food Hygiene Association (2008) Guidelines for Food Hygiene Inspection Microorganism. Japan, Tokyo. pp: 345-348.
- Barrow GI, Feltham RKA (1993) Cowan and Steel’s manual for the identification of medical bacteria. (3rdedn.) Cambridge University Press, Cambridge.
- Matsuyama H, Katoh H, Ohkushi T, Satoh A, Kawahara K, et al. (2008) Sphingobacterium kitahiroshimense sp. nov., isolated from soil. Int J Syst Evol Microbiol 58: 1576-1579.
- Altschul SF, Madden TF, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402.
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680.
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95-98.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874.
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425.
- Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111-120.
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783-791.
- Andrikopoulos S, Blair AR, Deluca N, Fam BC, Proietto J (2008) Evaluating the glucose tolerance test in mice. Am J Physiol Endocrinol Metab 295: E1323-E1332.
- Barrow GI, Feltham RKA (1993) Cowan and Steel’s manual for the identification of medical bacteria. (3rdedn.). Cambridge University Press, Cambridge.
- Brady CL, Goszczynska T, Venter SN, Cleenwerck I, De Vos P, et al. (2011) Pantoea allii sp. nov., isolated from onion plants and seed. Int J Syst Evol Microbiol 61: 932-937.
- Grimont PAD, Grimont F (2005) Genus XII. Enterobacter Hormaeche and Edwards. Pp: 661-669. Springer, New York.
- Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, et al. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122-1126.
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959-962.
- Weyer C, Funahashi T, Tanaka S, Hotta K, Matsuzawa Y, et al. (2001) Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab 86: 1930-1935.
- Bagarolli RA, Tobar N, Oliveira AG, Araújo TG, Carvalho BM, et al. (2017) Probiotics modulate gut microbiota and improve insulin sensitivity in DIO mice. J Nutr Biochem 50: 16-25.
- Frazier TH, DiBaise JK, McClain CJ (2011) Gut microbiota, intestinal permeability, obesity-induced inflammation, and liver injury. JPEN J Parenter Enteral Nutr 35: 14S-20S.
- Cani PD, Delzenne NM, Amar J, Burcelin R. (2008) Role of gut microflora in the development of obesity and insulin resistance following high-fat diet feeding. Pathol Biol (Paris) 56: 305-309.
- Tilg H, Kaser A (2011) Gut microbiome, obesity, and metabolic dysfunction. J Clin Invest 121: 2126-2132.
Copyright: © 2018 Yamanouchi K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.