
Journal of Clinical and Cellular Immunology
Open Access
ISSN: 2155-9899

ISSN: 2155-9899
Research Article - (2020)Volume 11, Issue 6
The immune responses underlying the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remain unclear. To help understand the pathology of coronavirus disease 2019 (COVID-19) pandemics, public data were analyzed and the expression of PDCD1 (encoding PD-1) and CD274 (encoding PD-L1) in T cells and macrophages were identified to correlate positively with COVID-19 severity.
COVID-19; Macrophages; PDCD1; CD274; T cells
mDCs: myeloid Dendritic Cells; pDCs: plasmacytoid Dendritic Cells; ICMs: Inhibitory Checkpoint Molecules; SCMs: Stimulatory Checkpoint Molecules; IL-6: Interleukin 6; BALF: Bronchoalveolar Lavage Fluid
COVID-19 has led to the global pandemic and infected millions of people worldwide [1]. It’s urgent to better understand the pathophysiology of the infection caused by SARS-CoV-2 [2-4]. The PD-1/PD-L1 signaling plays an essential role not only in regulating tumor immune responses but also in balancing homeostasis and tolerance in virus infection [5], but its role in COVID-19 is currently unclear. Thus, it is necessary to investigate how PD-1/ PD-L1 signaling works during COVID-19 progress in order to deal with it.
Single-cell immune profiling was explored in COVID-19 patients using the public data, which is essential for understanding the potential mechanisms underlying COVID-19 pathogenesis.
Data sets and processing
We downloaded the gene expression matrix from GEO database (GSE145926 and GSE128033). This dataset contained 63734 cells from four healthy people, three patients with moderate infection by bilateral pneumonia and seven patients with severe infection. The detailed clinical information of patients can be obtained in the reference [6]. Single cell clustering and cell annotation was performed as previously described [6].
Statistical analysis
The Student’s t-test (t.test in R 3.5.1) was used across three experiment designs. *P<0.05, **P<0.005, ***P<0.001. Values are presented as mean Standard Deviation (SD). Error bars represented the SD.
Single-cell analysis revealed distinct immune cell subsets
Publicly available data of bronchoalveolar cells from three moderate (M1-M3) and six severe (S1-S6) COVID-19 patients, and four healthy controls (HC1-HC4) were collected for analysis (66630 cells, Table S1) [6]. 31 clusters were identified by classical signature genes according to the reference (Figure 1) [6]. CD68-Macrophage (0, 1, 2, 3, 5, 6, 7, 8, 9, 11, 12, 15, 19, 21, 24, 30) dominated among these populations, followed by CD3D-T cell (4, 10, 16).
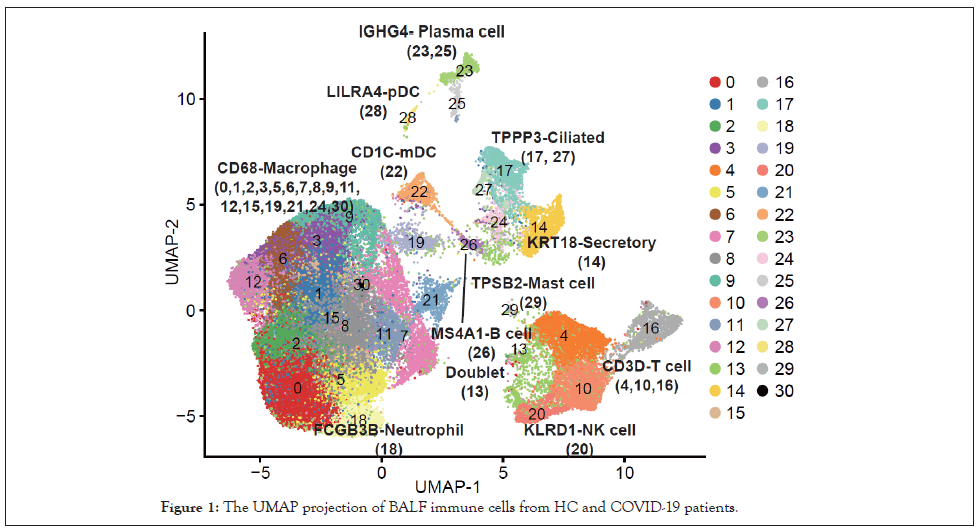
Figure 1: The UMAP projection of BALF immune cells from HC and COVID-19 patients.
Various expression patterns of PDCD1 and CD274 in different cell subpopulations
Expression of PDCD1 was first analyzed at the patient group level in different cell subpopulations, and four trends of their expression dynamics were observed (Figure 2). PDCD1 expression was gradually elevated in T cell, B cell, myeloid Dendritic Cells (mDCs), and macrophages from HC to mild cases then to severe patients (Figure 2A). In the 2nd trend, PDCD1 expression was specifically increased in plasma cells and epithelial cells in severe patients but not in mild patients (Figure 2B). For the 3rd trend, PDCD1 expression was upregulated in mild patients but slightly reduced in severe patients in NK and plasmacytoid Dendritic Cells (pDCs) (Figure 2C). No expression of PDCD1 was detected in mast cells and neutrophils in the 4th trend (Figure 2D).
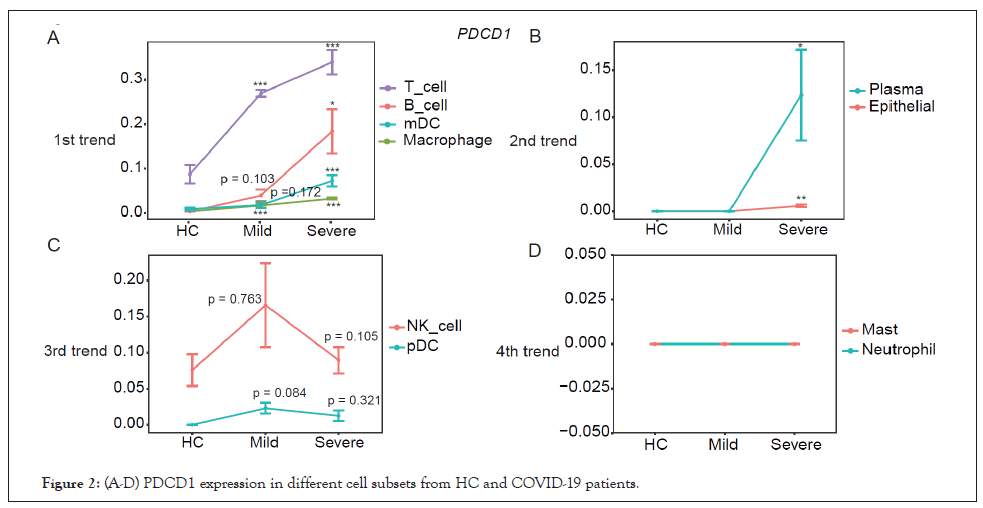
Figure 2: (A-D) PDCD1 expression in different cell subsets from HC and COVID-19 patients.
The CD274 expression in macrophages, mast cells, pDC, and T cells (1st trend) correlated well with COVID-19 severity (Figure 3A) and was specifically increased in plasma cells of severe patients (2nd trend) (Figure 3B). However, in neutrophil, mDC, B cells and NK cells, the expression of CD274 was upregulated in patients with mild COVID-19, then decreased in severe patients (Figure 3C). In contrary to the 3rd trend, the expression of CD274 was decreased in mild patients but increased in severe patients (Figure 3D).
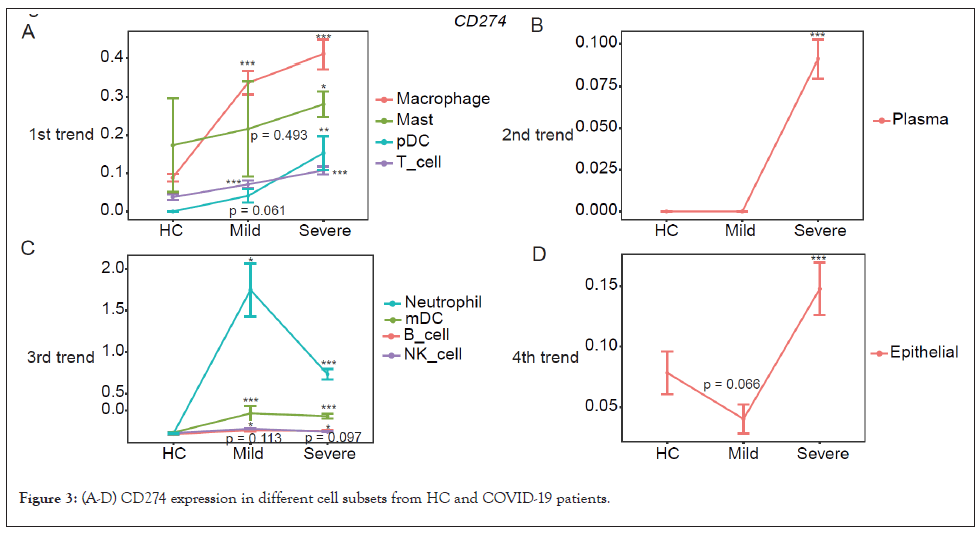
Figure 3: (A-D) CD274 expression in different cell subsets from HC and COVID-19 patients.
When analyzed at the individual level, expression of PDCD1 and CD274 was also elevated in mild and severe patients (Figures 4A and 4B). Overall, PDCD1 expression in T cells, B cells, mDCs, and macrophages and CD274 expression in macrophages, mast cells, pDC, and T cells correlated well with COVID-19 severity. Furthermore, PDCD1 and CD274 expression was specifically increased in epithelial and plasma cells of severe patients.
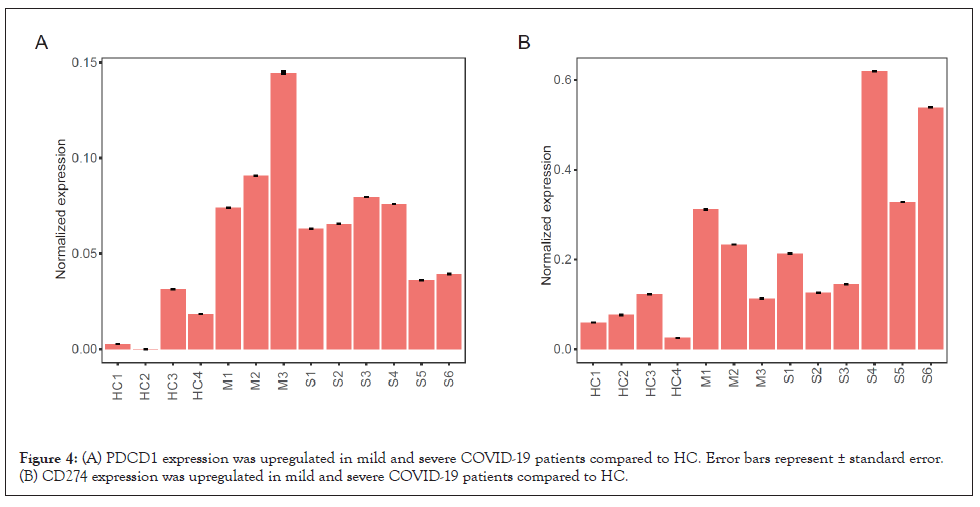
Figure 4: (A) PDCD1 expression was upregulated in mild and severe COVID-19 patients compared to HC. Error bars represent ± standard error. (B) CD274 expression was upregulated in mild and severe COVID-19 patients compared to HC.
The expression of STAT1 was increased in patients with mild and severe COVID-19
Inflammatory signaling participates in modulating PD-L1 expression, particularly, STAT1, which can be activated by IFNγ or Interleukin 6 (IL-6), is a crucial regulator for PD-L1 expression [7,8]. Furthermore, plasma IFNγ level [3] and the IL-6 level in Bronchoalveolar Lavage Fluid (BALF) [6] were reported to be increased in COVID-19 patients. Consistently, STAT1 was found upregulated in both mild and severe patients (Figure 5), suggesting increased CD274 expression might at least partly resulting from increased STAT1 level in COVID-19 patients.
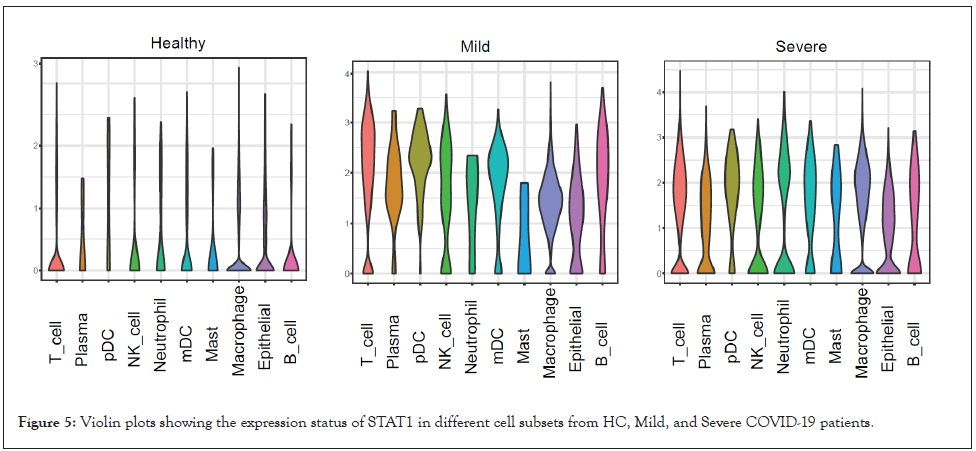
Figure 5: Violin plots showing the expression status of STAT1 in different cell subsets from HC, Mild, and Severe COVID-19 patients.
The landscape of immune checkpoint molecules in COVID-19 patients
To further elucidate the immune checkpoint landscape of COVID-19 patients, expression of classical inhibitory and stimulatory checkpoint molecules was assessed. For Inhibitory Checkpoint Molecules (ICMs), expression of CD160, CD244, PDCD1, BTLA, TIGIT, LAG3, KLRG1, and ADORA2A were increased in mild patients compared to HC and severe patients while expression of CTLA4, HAVCR2, IDO1, and CD276 were highest in severe patients (Figure 6A). Regarding Stimulatory Checkpoint Molecules (SCMs), expression of TNFRSF9, CD28, ICOS, and CD27 was elevated in mild patients in comparison to HC and severe patients while expression of TNFRSF18 and CD40 were highest in severe patients (Figure 6B).
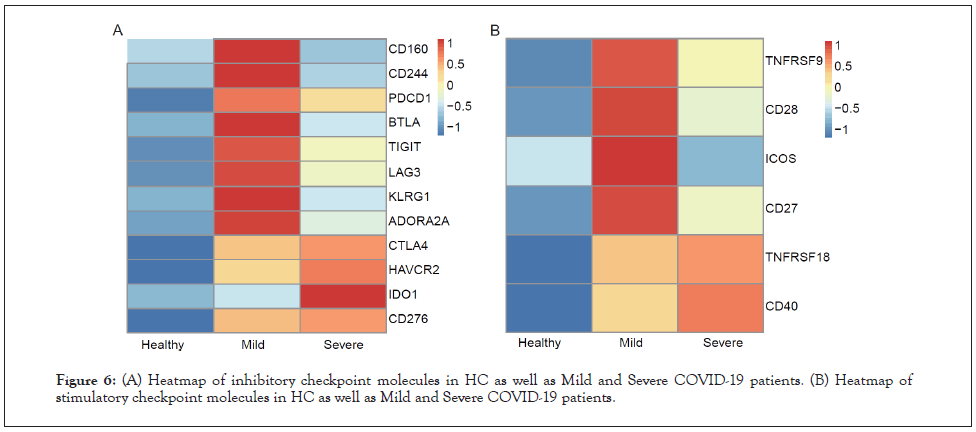
Figure 6: (A) Heatmap of inhibitory checkpoint molecules in HC as well as Mild and Severe COVID-19 patients. (B) Heatmap of stimulatory checkpoint molecules in HC as well as Mild and Severe COVID-19 patients.
When analyzed at the cell subpopulation level, unique expression patterns of ICMs and SCMs were demonstrated in each cell subpopulation (Figures 7 and 8). Expression patterns of ICMs were different between mild and severe patients (Figure 7). Most categories and the highest intensity of ICMs were enriched in severe patients, except in T cells and NK cells. The results indicated that the immune cells might be more exhausted in severe patients with COVID-19 compared to healthy people and even mild patients. In terms of SCMs, macrophages, mast cells, and pDC of severe patients got more enriched than that of mild patients and healthy donors (Figure 8). In T cell population, more ICMs and SCMs were enriched in patients with mild and severe COVID-19 than that of healthy donors, but there was no dramatic difference between mild and severe patients themselves (Figures 7 and 8).
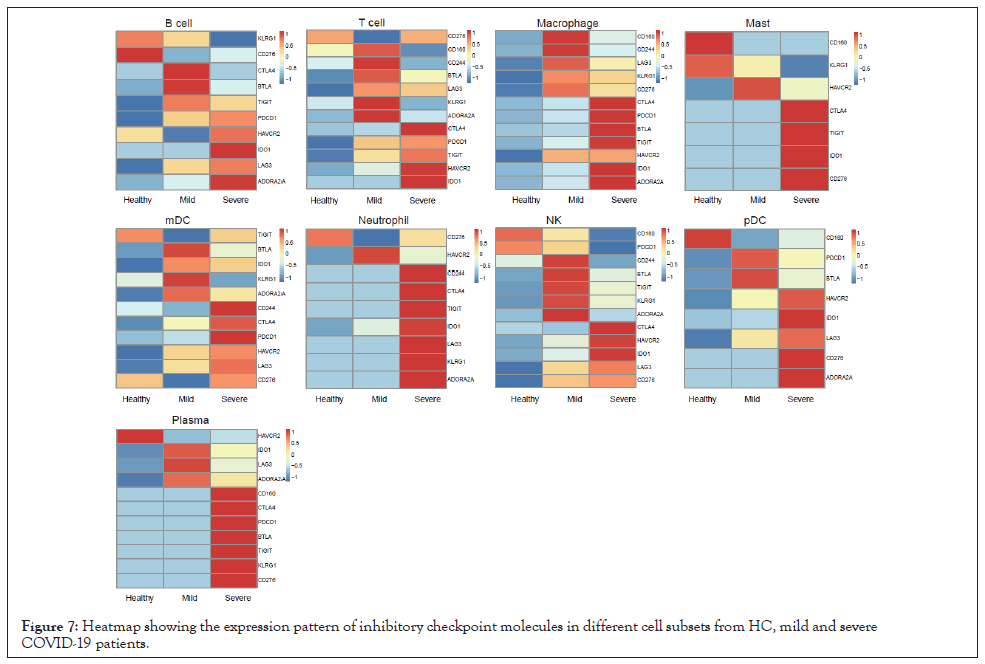
Figure 7: Heatmap showing the expression pattern of inhibitory checkpoint molecules in different cell subsets from HC, mild and severe COVID-19 patients.
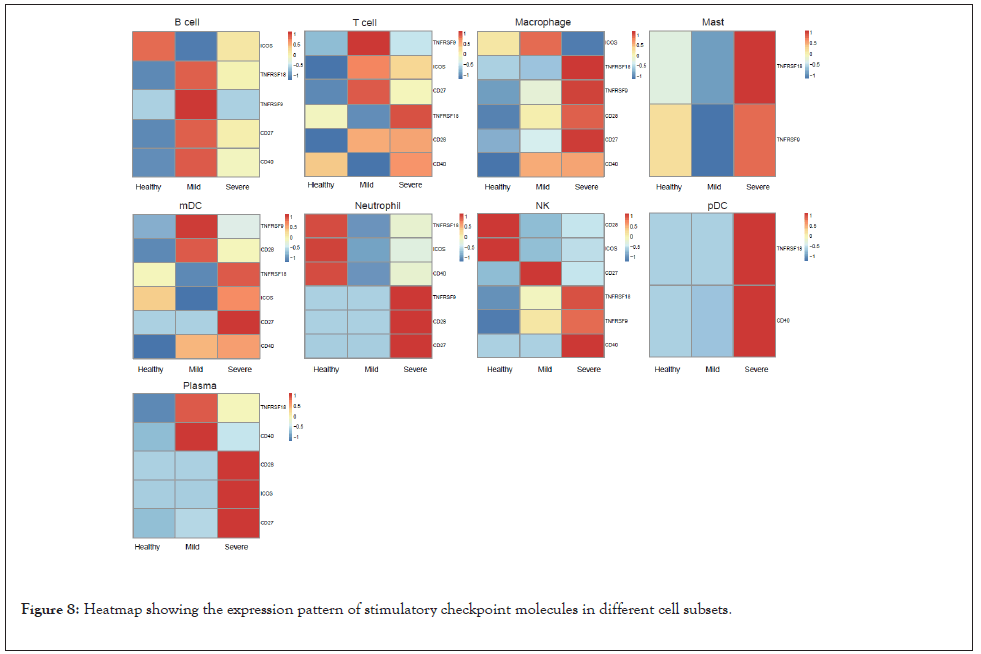
Figure 8: Heatmap showing the expression pattern of stimulatory checkpoint molecules in different cell subsets.
Since COVID-19 is pandemic and threatening thousands of people’s life, it is urgent and essential to investigate the molecular mechanism of the immune pathogenesis of the disease. Compared to healthy controls, PDCD1 expression in T cells, B cells, mDCs, and macrophages (Figure 2) and CD274 expression in macrophages, mast cells, pDCs, and T cells (Figure 3) were upregulated in COVID-19 patients, and correlated well with COVID-19 severity. Moreover, expression of PDCD1 and CD274 was specifically increased in plasma cells of severe patients (Figures 2 and 3), which could serve as a biomarker for prognosing the severity of COVID-19. Many clinical trials for treating COVID-19 are ongoing. Among them, one clinical trial uses PD-1 monoclonal antibody to block PD-1 in COVID-19 patients (NCT04268537). Based on our results, PDCD1 expression was dramatically upregulated in T cells and macrophages especially in severe patients (Figure 2) and its blockade would further increase the secretion of multiple proinflammatory cytokines, which will enhance the cytokine release syndrome reported in COVID-19 patients and possibly associated with disease severity [2,3], leading to further tissue damage or even more death especially in severe COVID-19 patients [9,10]. A current study supports that checkpoint inhibitor immunotherapy is risky for severe outcomes in SARS-CoV-2-infected cancer patients, though these patients were treated with Immune Checkpoint Inhibitors (ICI) before SARS-CoV-2 infection [11].
Our research provides valuable information about COVID-19 and its severity correlated well with the expression of PDCD1 and CD274.
The authors declare that they have no competing interests.
Gao Q supervised the project, designed the bioinformatic analysis, wrote and revised the manuscript. Liu S. performed the bioinformatic analysis. Ding R dealt with the data. Chen H, Dong X, Xie J, Li Y, Chen L, and Liu H helped with the data management.
We sincerely thank the support provided by China National GeneBank and this research was supported by the Guangdong Enterprise Key Laboratory of Human Disease Genomics (2020B1212070028), and Science, Technology and Innovation Commission of Shenzhen Municipality (JSGG20180508152912700).
Citation: Gao Q, Liu S, Ding R, Chen H, Dong X, Xie J, et al. (2020) The Expression of PDCD1 and CD274 in T cells and Macrophages Correlated Positively with COVID-19 Severity. J Clin Cell Immunol. 11:603. doi: 10.35248/2155-9899.20.11:603.
Received: 14-Sep-2020 Accepted: 28-Sep-2020 Published: 05-Oct-2020 , DOI: 10.35248/2155-9899.20.11.603
Copyright: © 2020 Gao Q, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Sources of funding : We sincerely thank the support provided by China National GeneBank and this research was supported by the Guangdong Enterprise Key Laboratory of Human Disease Genomics (2020B1212070028), and Science, Technology and Innovation Commission of Shenzhen Municipality (JSGG20180508152912700).