Advancements in Genetic Engineering
Open Access
ISSN: 2169-0111
ISSN: 2169-0111
Research Article - (2023)Volume 12, Issue 2
Fusion genes had been thought to be somatic and cause cancer, including Acute Myeloid Leukemia (AML). Validating highly-recurrent fusion genes in healthy samples compelled us to systematically study Hereditary Fusion Genes (HFGs). Here, we used the previously-curated 1180 HFGs from Monozygotic (MZ) twins to analyze total fusion transcripts from AML patients and identified 926 (78.5%) HFGs overlapped with AML. We selected 242 HFGs ranging from 10% to 83.3% to perform comparative analysis and showed that 239 HFGs were significantly higher than their counterparts of Genotype-Tissue Expression (GTEx) blood samples and associated with AML inheritances. A 5’-gene and 3’-gene were fused with multiple 3’-genes and 5’-genes, respectively, to form biological networks and result in potential hyper susceptibility to somatic genetic and environmental abnormalities. Many highly-recurrent HFGs were also observed in Multiple Myeloma (MM) and MZ twins, suggesting that complex traits and diseases shared common hereditary factors. This study shed the first light on HFGs as most “inherited” genetic factors and provided potential therapeutic targets, leading to paradigm shifts in cancer and disease diagnosis and research
Hereditary; Epigenetic; Fusion genes; RNA-seq; Genetics; Genomics; Leukemia
Acute Myeloid Leukemia (AML) is a type of blood cancer characterized by uncontrolled clonal expansion of hematopoietic progenitor cells. Cytogenetic profiles and molecular screening found that the acquired recurrent genomic abnormalities aggravated the cell’s malignant growth [1]. Advances in nextgeneration genome-wide sequencing and RNA Sequencing (RNA-Seq) have made it possible to identify gene mutations [2] and genomic abnormalities [3], resulting in fusion genes [4]. Even though familial clustering of AML cases had been known for more than five decades, several syndromes were shown to be genetic predisposition syndromes to AML development [5]. Although germline mutations associated with leukemia were discovered in more than 20 genes, they could explain only small percentages of these genetic predispositions. The germline genetic lesion in a significant fraction of these families remained unknown [5-7].
We analyzed large RNA-Seq datasets and identified hundreds of thousands of fusion genes, from which KANSARL (KANSL1- ARL17A) was validated as the first predisposition fusion gene specific to 29% of populations of European ancestry [8]. TPM4- KLF2, detected in 92.2% of Multiple Myeloma (MM) patients and 1% of GTEx blood samples, had been detected in all five healthy bone marrows [9]. ADAMTSL3-SH3GL3 had been detected in 100% of non-neurological controls and 87.5% of Alzheimer’s disease healthy controls [10]. These fusion gene data raised doubts that fusion genes were generated via somatic genomic abnormalities and compelled us to use the Monozygotic (MZ) twin genetic model to systematically study Hereditary Fusion Genes (HFGs) and discover 1180 HFGs [11]. HFGs were defined as the fusion genes offspring inherited from their parents, excluding read-through transcripts, which were named epigenetic fusion genes [11]. In this report, we used the curated HFGs to analyze fusion transcripts uncovered from 390 AML patients’ RNA-Seq and identified 926 potential HFGs, 242 of which were classified as HFGs ranging from 10% to 88.3%. We characterized these 242 HFGs and indicated that HFGs were the dominant genetic factors associated with AML.
Materials
Human Acute Myeloid Leukemia (AML) RNA-Seq dataset: RNA-Seq data of the Leucegene AML project [12] (GEO: GSE67040) were downloaded from NCBI. We identified heparinized blood samples of 391 unique AML patients. We removed one sample with bad-quality RNA-Seq from this study. Hence, this dataset had 390 unique AML patients.
The Genotype-Tissue Expression (GTEx) blood RNA-Seq: We selected GTEx blood samples as a control to evaluate the results better. We downloaded the GTEx blood RNA-Seq (dbGapaccession: phs000424.v7.p2), from which we identified 462 healthy blood samples.
Methods
Identification of fusion transcripts by Splicing Codes Identify Fusion (SCIF) transcripts: Splicing Codes Identify Fusion (SCIF) transcripts was developed in C and Perl program languages. The algorithm used by SCIF has been described in detail previously [8]. More details are described in Supporting Materials.
Identifying Hereditary Fusion Genes (HFGs) in Acute Myeloid Leukemia (AML): To discover HFGs in AML, we first used SCIF to analyze RNA-Seq data of the Leucegene AML project (GEO: GSE67040) at the default parameters and discovered 1.1 million fusion transcripts. Then, we determined the quality of uncovered fusion transcripts by manually inspecting some fusion transcripts from KANSARL and epigenetic (read-through) fusion genes (EFGs). Then, we compared these raw fusion transcripts with those from 37 pairs of Monozygotic (MZ) twins to ensure that both data were clear and relatively comparable. After inspecting, we used 1180 HFGs discovered and curated from the previous study of the MZ twins [11] to analyze the total fusion transcripts from 390 ALM patients. We used 5' 20 bp and 3' 20 bp fusion junction sequences to discover the fusion transcripts with identical 5' and 3' fusion junction sequences. If an RNA-Seq read had identical fusion junctions, this fusion transcript was thought to be from HFGs. Furthermore, we removed 67 fusion transcripts that overlapped with fusion transcripts of MZ HFGs, with too many alternative fusion gene IDs and only one isoform. We identified a total of 926 potential HFGs. Since we could not distinguish the HFGs with low recurrent frequencies from somatic fusion genes, we set the cutoff of recurrent frequency at 10%. Only potential HFGs with recurrent frequencies of ≥ 10% were treated as HFGs in this study. We obtained 242 HFGs.
Recurrent Frequencies (RFs): The Recurrent Frequency (RF) was defined as the number of HFG-positive individuals divided by the total number of samples used in a study. To rule out the potential effects of different experiments and protocols, if a sample was found to have one fusion junction of an HFG, this sample was treated as HFG-positive regardless of the copy numbers of this HFG.
Generation of heat maps by Morpheus: We first produced files of 390 AML patients possessing 242 HFGs with recurrent frequencies of ≥ 10% to generate heat maps. Then we used Morpheus (https://software.broadinstitute.org/morpheus/) from Broad Institute. We first used K-means clustering with Euclidean distance and three members to cluster 242 HFGs and 390 AML patients. Then, we further clustered the HFGs and patients by hierarchical clustering. The heat map was saved as a PDF file.
Identification and characterization of Hereditary Fusion Genes (HFGs) associated with acute myeloid leukemia
We downloaded RNA-Seq data of the Leucegene AML project [2](GEO: GSE67040) from NCBI, which contained heparinized blood samples of 390 AML patients. We used SCIF (Splicing Codes Identify Fusion Genes) to analyze RNA-Seq data and identified 1,010,000 fusion transcripts, 114,000 of which were detected in 2 to 325 AML patients. Alternative splicing and repetitive sequences caused false positives, which were 1%-2% [8]. Since the random genomic alteration rate was 3.6 × 10-2 [13,14], HFG frequencies in a population would be 20 folds higher than that of random genomic alterations [11] and hence distinguish somatic abnormalities. We used 1180 HFGs previously discovered from MZ twins to search AML fusion transcripts [11]. To avoid confusion between low-recurrent HFGs and fusion genes created by somatic abnormalities, we set 10% Recurrent Frequency (RF) as the cutoff. Therefore, unless specified, HFGs with recurrent frequencies of ≥ 10% were treated as HFGs or potential HFGs. We uncovered 638 potential HFGs and 242 HFGs ranging from 10% to 83.3%, the average of which was 24%. 242 HFGs counted for 23.8% of 1027 fusion genes with RFs of ≥ 10% and were significantly larger than the germline mutations identified [6], suggesting that HFGs were the dominant genetic factors. To better evaluate the results, GTEx 462 healthy blood samples were selected as control (dbGapaccession: phs000424.v7.p2). Similarly, 1180 MZ twin HFGs were used to analyze fusion transcripts from GTEx and uncovered 383 potential HFGs ranging from 0.2% to 37.4%, the average of which was 1.7%. Then, 42.6% of 383 GTEx HFGs overlapped with 242 AML HFGs and counted for 67.1%, supporting that HFGs were conserved and widespread [11] (Figure 1).
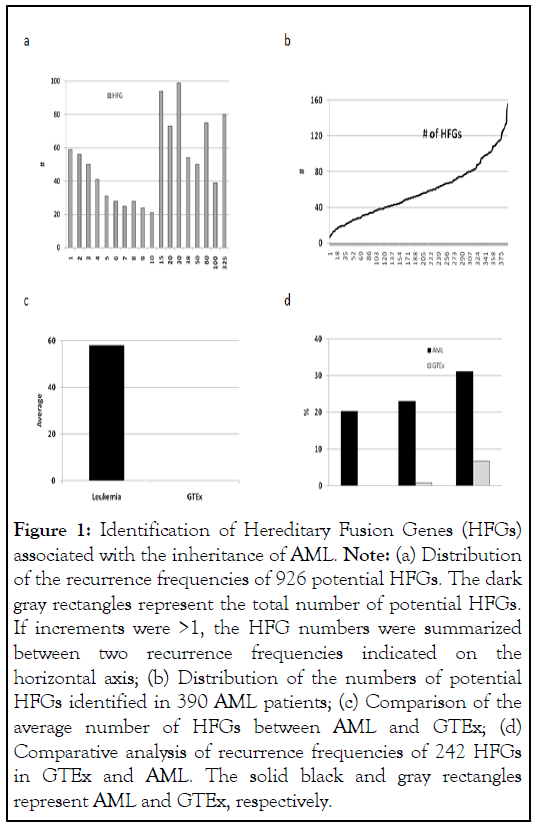
Figure 1: Identification of Hereditary Fusion Genes (HFGs) associated with the inheritance of AML. Note: (a) Distribution of the recurrence frequencies of 926 potential HFGs. The dark gray rectangles represent the total number of potential HFGs. If increments were >1, the HFG numbers were summarized between two recurrence frequencies indicated on the horizontal axis; (b) Distribution of the numbers of potential HFGs identified in 390 AML patients; (c) Comparison of the average number of HFGs between AML and GTEx; (d) Comparative analysis of recurrence frequencies of 242 HFGs in GTEx and AML. The solid black and gray rectangles represent AML and GTEx, respectively.
We identified 926 (78.5%) potential HFGs ranging from 1 to 325 AML patients (Figure 1a), suggesting that most HFGs had negative impacts. Figure 1b showed that AML patients had from 6 to 156 HFGs, the average of which was 58 and 944-folds higher than the GTEx one (Figure 1c), suggesting that these 242 HFGs were infrequent in healthy populations and associated with AML. Figure 1d showed that as the RFs of GTEx HFGs increased, so did the AML counterparts, suggesting that most HFGs were under selection pressures. The statistical analysis showed that the RFs of 239 AML HFGs were significantly higher than the GTEx counterparts, suggesting that these 239 HFGs were associated with AML. The recurrent frequencies of AML HFGs were 1.5 to 292 folds of the GTEx blood counterparts. The highest difference between AML and GTEx was SYNCRIPEEF1A1, detected in 63.3% of AML patients and 0% of GTEx blood samples and encoded a putative intact eukaryotic translation elongation factor 1 α1, suggesting that altering EEF1A1 gene expression played a role in AML.
Characterizations of Hereditary Fusion Genes (HFGs) associated with Acute Myeloid Leukemia
To characterize HFGs, we sorted 5' and 3' HFG gene IDs, respectively (Figure 2). Figure 2a showed that seven 5’-genes were fused with ≥ 5 3’-genes and three folds of the 5’-genes’ average, suggesting 5’-genes were not randomly distributed. OAZ1 was the most recurrent 5’-fused gene and fused with 24 different 3'-genes. Out of 11 fusion sites observed, all main isoforms of these 24 5'-OAZ1-fused genes used the first OAZ1 exon (NM_004152.2). OAZ1 provided promoter region, 5’- UTR, and ATG depending on 3'-genes. Figure 2b showed that AML patients possessed up to 21 different OAZ1-fused HFGs, the average of which was 5.4. Numerous 5’-OAZ1-fused HFGs in an individual indicated that identical regulatory factors controlled these HFGs to form a natural one-to-many work. In contrast, Figure 2b showed that 52.5% of GTEx individuals had no single copy of 5’-OAZ1-fused HFGs, the average of which was 0.48 and 11-fold less than the former. Consequently, a single environmental factor or genetic alteration could affect all genes under this network and quickly initiate and develop AML.
Figure 2: Characterization of 242 HFGs associated with AML inheritance. Note: (a) The list of 5'-genes fused with ≥ 5 3'- fused genes; (b) The comparative analysis of 5'-OAZ1-fused HFGs among 390 AML patients and 462 GTEx individuals; (c) The list of 3'-genes fused 24 with ≥ 5 5'-fused genes; (d) The comparative analysis of 5'-EEF1A1-fused HFGs among 390 AML patients and GTEx individuals. Solid black and gray rectangles represent AML and GTEx, respectively.
Figure 2c showed that eleven 3'-genes were fused with ≥ 5 5'- genes. The most recurrent 3'-gene was EEF1A1 and was fused with 17 different 5'-genes. The most recurrent isoforms of all 3'- EEF1A1-fused HFGs used the 5' splice site of the second EEF1A1 exon (NM_001402.5), located at 5' UTR of EEF1A1. All 5' genes provided their regulatory and 5’UTR sequences, conferring only regulatory machinery. All seventeen 3'-EEF1A1- fused HFGs encoded intact eukaryotic translation elongation factor 1 α1, whose overexpression inhibited p53 and p73- dependent apoptosis and chemotherapy sensitivity in cervical carcinoma cells [15]. Figure 2d showed that the numbers of 5’- genes fused with EEF1A1 ranged from zero to 17, the average of which was 6.3. One individual's maximum 5' genes were 17 and formed a many-to-one network, which disrupted EEF1A1 expression. In contrast, Figure 2d showed that 83.3% of GTEx individuals had no EEF1A1-fused counterparts, and the average was 0.013. The former was 484-fold higher, suggesting that 3'- EEF1A1-involved HFGs were associated with AML.
Figures 2a and 2c showed that Actin Beta (ACTB) and DEADbox helicase 5(DDX5) encoded beta-actin and DEAD-box helicase 5, respectively, and were fused with 5'-and 3'-genes to form both one-to-many and many-to-one networks. A 5’-ACTB and 3’-ACTB were fused with six different 3’-genes and twelve 5’-genes to form six 5’-ACTB HFGs and twelve 3’-ACTB HFGs. The 5’-ACTB regulatory sequences [16] dramatically enhanced six HFGs overexpression. On the other hand, 12 5’-genes were fused with 3’-ACTB to form 12 3’-ACTB-fused HFGs, eight expressing truncated or hybrid proteins. 3’-ACTB-fused HFGs and their recurrent frequencies were more remarkable than ACTB gene mutations [17] and dramatically increased AML complexities. Therefore, cells under stress underwent natural selection to promote somatic abnormalities and mutations and quickly result in genetically and phenotypically heterogeneous clonal hematopoietic progenitors [18].
Characterization of hereditary fusion gene genotypes of acute myeloid leukemia patients
To discover the characteristics of HFG genotypes of AML patients, we used Morpheus (https://software.broadinstitute.org/ morpheus/) to cluster these 242 HFGs. Figure 3 showed that HFGs were clustered into three groups-the highly-recurrent, moderately-recurrent, and sparsely-recurrent HFGs, which consisted of 11.5%, 38.3%, and 50.2%and were consistent with HFG RFs (Figure 1a).
Figure 3: Heatmap of 242 HFGs among 390 AML patients generated by Morpheus. Horizontal light green: Highlyrecurrent; Blue: Moderately-recurrent; Yellow rectangles: Sparsely-recurrent HFGs. While Vertical blue, yellow, and light green rectangles represent patients with highly abundant, moderately abundant, and sparsely-abundant HFGs, respectively.
There were 28 HFGs among the highly-recurrent HFGs ranging from 40% to 83.3%, the top eleven HFGs of which Table 1 showed. A quarter of them was 3'-EEF1A1-fused HFGs, indicating that disrupting EEF1A1 expression played essential roles in AML. TPM4-KLF2 [19], OAZ1-KLF16 [20], and YPEL5- FOSL2 were HFGs that disrupted transcription factors. TPM4- KLF2 was first reported in acute lymphoblastic leukemia [19]. It was later found in 30% of AML samples and all three normal bone marrow samples [21], suggesting that defected transcription factors played essential roles in leukemia initiation and development.
| 5' Gene | 3' Gene | AML(%) | MM(%) | GTEx(%) |
|---|---|---|---|---|
| TRNAN35 | FAM91A3P | 83.3 | 58.2 | 13.1 |
| C9orf100 | DENND1C | 74.6 | 17.7 | 40.5 |
| TPM4 | KLF2 | 74.1 | 92.2 | 0.9 |
| OAZ1 | KLF16 | 67.9 | 12.8 | 5.2 |
| PTP4A1 | EEF1A1 | 67.7 | 25.2 | 2.8 |
| YWHAE | CRK | 67.7 | 14.6 | 1.4 |
| SYNCRIP | EEF1A1 | 63.3 | 12.2 | 0 |
| BACH1 | MECP2 | 56.7 | 15.8 | 4.4 |
| B2M | CHD2 | 49.5 | 14 | 0.9 |
| DDX5 | EEF1A1 | 49.5 | 17.2 | 0 |
| YPEL5 | FOSL2 | 49.5 | 10 | 0 |
Table 1: Recurrent frequency comparison of eleven HFGs between 390 AML and 727 Multiple Myeloma (MM) patients. 462 GTEx blood samples were used as control.
It has been reported that Multiple Myeloma (MM) patients had an 11.51-fold increased risk of AML [22], hinting that both MM and AML had common genetic infrastructures. To examine this idea, we compared these eleven HFGs between AML and MM to investigate if they shared HFGs. Table 1 showed that the eleven AML HFGs were also found in MM patients ranging from 10% to 92.2% and confirmed that some AML and MM patients shared common genetic factors. The most common HFGs between AML and MM were TRNAN35-FAM91A3P and TPM4- KLF2, encoding non-coding fusion RNA and a tropomyosin 4 and kruppel-like factor 2 fusion protein. Both HFGs were detected in 56.4% and 92.2% of 727 MM patients [9]. Supplementary Table 2 showed that eight 5'-genes were fused with 3'-KLF2 to form 3’-fused HFGs ranging from 11.3% to 74.1%. DNA analysis showed that ACTB-KLF2, AKAP8- KLF2, B2M-KLF2, and PTBP1-KLF2 resulted in frame shifts to produce ORFs having similarities with kruppel-like transcription factors. TPM4-KLF2, ELL-KLF2, and EPS15L1- KLF2 were in-frame HFGs and produced fusion proteins. Figure 1 showed that 341 (87.4%) AML patients had from one to eight 3'-KLF2-fused HFGs, the average of which was 2.7 and 63-folds of the GTEx counterpart. Only 4% of the GTEx populations possessed one of the eight 3'-KLF2-fused HFGs, confirming that disrupting KLF2 genes played critical roles in AML initiation and development. Supplemantary Table 3 showed that three additional 5'-and six 3'-KLF-fused HFGs altered kruppel-like factor expression. These 5' and 3'- KLF-fused HFGs directly interfered with gene expression patterns and promoted somatic genomic alterations in the later developmental stages.
Figure 3 showed that based on individual HFG genotypes, AML patients were clustered into three groups: AML patients with abundant, moderate, and sparse HFGs, which consisted of 18.5%, 38.2%, and 43.3%. AML patients with abundant HFGs had all types of HFGs, from sparsely recurrent HFGs to highlyrecurrent HFGs, and suggested that many HFGs functioned as groups. To understand the nature of the abundant HFG AML, we used Synthesis Of Cytochrome C Oxidase 2 (SCO2)-fused HFGs as examples generated by local SCO2 amplification associated with MZ twin inheritance [11]. They included PIM3- SCO2, PLXNB-SCO2, PPP6R2-SCO2, and TRABD-SCO2 and were detected in 67.6%, 47.3%, 36.5%, and 20.3%, respectively [11]. They were observed in 29.2%, 32.6%, 37.9%, and 12.8% of AML patients. Figure 2 showed that 30.8% of AML patients possessed ≥ 2 SCO2-fused HFGs, and 19 AML patients had all four SCO2-fused HFGs. Therefore, these closely-linked SCO2- fused HFGs functioned as groups and were passed to offspring. All these data showed that AML patients had complex HFG genotypes, constituting the AML genetic infrastructures.
As Figure 3 showed that 43.3% of AML patients were HFGsparse, these patients did not mean that they were HFGs-sparse for four reasons. First, 242 FHGs were only 23.8% of 1027 fusion genes with RFs of ≥ 10%. Since ≥ 80% of these highlyrecurrent fusion genes were predicted to be potentially inherited [9], more HFGs required further validation. Second, since there were 683 potential HFGs, these potential HFGs added AML genetic knowledge. Third, the curated HFGs were only a tiny portion of 5×109 [11]. When HFGs increased, more HFGs would be found in AML patients. Moreover, our approach avoided the identification of majorities of fusion genes generated by tandem duplications. This type of HFGs was identified due to their pseudogenes, DNA duplications, and homologs [11]. From traditional genetics and evolution [23,24], tandem gene duplications provided the most potent and direct self-support evidence that HFGs were the most critical genetic concepts for future genetics and genomics. This work just revealed the tip of the HFG iceberg and marked the dawn of the digitalization of genetics and genomics.
In this report, we have used curated 1180 HFGs to analyze the fusion genes uncovered from 390 Acute Myeloid Leukemia patients and uncovered 926 potential HFGs, from which 242 HFGs were identified. We found out that the numbers of HFGs and their recurrent frequencies were significantly larger than the gene mutations identified so far.
One of the most recurrent AML HFGs is TPM4-KLF2, first reported in acute lymphoblastic leukemia, which has been detected in Multiple Myeloma and Monozygotic (MZ) twins, suggesting as common “inherited” genetic factors. Our findings strongly suggest that pediatric cancer is a potentially complex genetic disease. In case if RNA-Seq of parents and children is performed with a family cancer history and other genetic disorders, we can identify their HFGs and discover who will likely have pediatric cancer. This is a simple procedure which furnishes our HFG databases and will save ten thousand children's lives.
We have expressed our profound appreciation to Ms. Xiaoyan Yang, Prof. Benoit Chabot, Prof. Jeff Xiwu Zhou, Prof. Shunbin Ning, Prof. Yanbin Zhang, and Prof. Yinxiong Li for their various contributions and support during the last two decades. We thank the support from the science and technology innovation Program of Hunan Province.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Fei L, Yang J, Zhuo N, Zhuo D (2023) The Hereditary Fusion Genes are Associated with the Inheritance of Acute Myeloid Leukemia. Advac Genet Eng. 12:214
Received: 20-Mar-2023, Manuscript No. MAGE-23-22280; Editor assigned: 24-Mar-2023, Pre QC No. MAGE-23-22280 (PQ); Reviewed: 07-Apr-2023, QC No. MAGE-23-22280; Revised: 14-Apr-2023, Manuscript No. MAGE-23-22280 (R); Published: 21-Apr-2023 , DOI: 10.35248/2169- 0111.23.12.214
Copyright: © 2023 Fei L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.