Journal of Probiotics & Health
Open Access
ISSN: 2329-8901
ISSN: 2329-8901
Research - (2019)Volume 7, Issue 2
The gut microbiota is the complex community of microorganisms that inhabit the human intestine. Gut microbes participate in many aspects of human physiology, including health and disease. Food ingredients, drugs and other environmental factors can affect the gut microbiota, with possible consequences on human health.
Progress in microbiome research has significantly stimulated and expanded the interest in technologies to study the potential of different products to modulate the gastro-intestinal ecosystem. In this context we have developed a method, called the i-screen, to evaluate the effects of compounds on the human gut microbiota. The i-screen is an in vitro system that allows the anaerobic cultivation of microorganisms obtained from fecal material, and therefore representative of the highly diverse colonic microbiota. By means of specific analyses, the effects of test compounds on the gut microbiota composition and metabolic activity can be assessed.
The i-screen has proven to be an effective and versatile experimental model of the gut microbiota, routinely applied to evaluate the effects of food ingredients and drugs. This system constitutes a valid contribution to product development and a starting point for a better understanding of the role of gut microbiota in host health.
Gut microbiota; In vitro model; Metabolism; Preclinical; Drug development
The human intestine is colonized by dense and diverse communities of microorganisms, collectively referred to as the gut microbiota. These communities are unique for each individual, and they also differ on the basis of geographic areas, age groups (e.g. babies, elderly) and health conditions (e.g. obese, IBD) [1,2]. Scientific and technological advances have greatly contributed to a better understanding of the role of these microbes in the human body. Nowadays it is accepted that the metabolism of the host and of its microbes are tightly connected: gut microbes are involved in many different aspects of human physiology, such as nutrition and immunity, and they play important roles in the maintenance of health and the development of disease [3].
Food and pharmaceutical compounds have a bidirectional interaction with the gut microbiota: metabolic and antimicrobial effects of these substances can induce specific changes in its composition and function; at the same time microbial metabolism is responsible for the transformation of pharmaceutical compounds and the breakdown of food ingredients [4]. Predicting the fate of compounds and their impact on the gut microbiota is not an easy task. The intestinal environment hosts many different microbial species, metabolically interconnected in ways that are often yet to be described and understood [5]. While it may be straightforward to test a compound on pure cultures or on simplified microbial communities, it is not always possible to extrapolate the effects observed or predicted in these systems to the complex ecosystem of the gut [6]. To better understand the gut microbiota and the many ways in which it impacts our responses to food intake, pharmaceuticals, environmental toxins and other microbes, it is important to develop methods that allow the study of effects of compounds while adequately mimicking in vivo conditions.
In this article we describe the i-screen, an in vitro pre-clinical screening platform that mimics the intestinal microbiota and enables testing of food and pharmaceutical compounds. Since its development over 10 years ago, the i-screen has been frequently applied for the development of functional food products and drugs, and it has been validated in various in vivo studies.
The i-screen is a 96-well plate for the incubation of intestinal microorganisms. The conditions in the i-screen (temperature, growth medium) mimic to the ones found in the proximal colon, and they allow to i) maintain a large microbial diversity, ii) maintain high microbial density, and iii) support the growth of anaerobic bacteria. Because of its 96-well plate setting, the i-screen is a medium- to high-throughput system, allowing to simultaneously testing a high number of samples. Figure 1 exemplifies the workflow of a typical i-screen experiment. This consists of four important components:
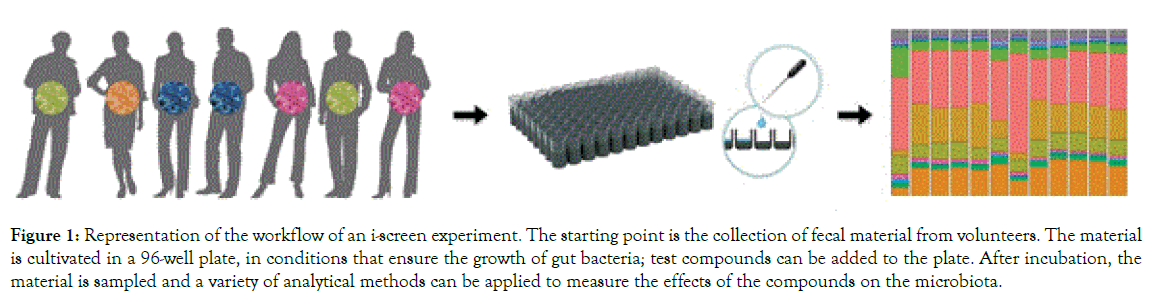
Figure 1. Representation of the workflow of an i-screen experiment. The starting point is the collection of fecal material from volunteers. The material is cultivated in a 96-well plate, in conditions that ensure the growth of gut bacteria; test compounds can be added to the plate. After incubation, the material is sampled and a variety of analytical methods can be applied to measure the effects of the compounds on the microbiota.
Cultivation medium
The chemical environment of the gut is characterized by the presence of partially digested and non-absorbed food compounds, cell debris, molecules excreted by the host, including mucins, and microbial metabolites. Throughout the large intestine, there is a gradient with decreasing levels of nutrients, as they are absorbed through the intestinal epithelium. During in vitro fermentation in the laboratory, the cultivation medium drives microbial metabolism. In order to support the growth of microorganisms originating from the colon, it is of key importance that the medium adequately simulates the conditions present in that environment. In the i-screen, this is achieved by using the standard ileal effluent medium (SIEM) as a growth medium. The composition of the SIEM is based on ileal effluent composition, which was originally developed for the TIM system [7] and has been optimized to obtain optimal growth in laboratory conditions. The medium composition can be fine-tuned to recreate conditions present in the large intestine of infants, elderly, and even farm animals.
Microbiota
The microbiota that is incubated in the i-screen is obtained from human fecal material. This is collected following a standardized protocol, handled in anaerobic conditions to ensure the survival of colonic microorganisms, and kept frozen at -80°C until use in an i-screen experiment. To account for inter-individual variability, i-screen incubations are routinely performed with pools of fecal material from different individuals. Reference pools are regularly assembled and characterized, representing specific population groups (healthy adults, infants, diseased groups, etc.) that provide a basis for standardized testing. Fecal samples obtained from individual donors can also easily be tested separately when a personalized approach is required. Furthermore, the starting microbiota can be supplemented with bacteria in order to study specific effects. For example, Clostridium difficile can be added to the microbiota to investigate the ecological effects of infection by a gastric pathogen, and to assess the impact of therapeutic interventions on the target bacteria as well as on the microbiota.
Cultivation conditions
Before starting an i-screen experiment, the fecal material is incubated overnight at 37°C in the appropriate cultivation medium and in anaerobic conditions, in order to prepare the microorganisms for the following incubation step. The material is then transferred to the 96-well plate where it will be incubated in the presence of specific test compounds. After 24 hours, samples are taken and analyzed with a variety of methods, depending on the research questions to address. Every experiment includes positive controls (compounds with well-known and characterized effect), negative controls (no added compounds), blanks (containing SIEM medium only, to control for contamination), and a number of additional technical controls, to ensures the reproducibility of i-screen experiments.
Read-outs
A variety of analytical methods can be applied to the material resulting from an i-screen experiment, in order to determine the effects of the compounds tested on the gut microbiota. These methods are summarized in Table 1. Changes in community composition and species abundance, in microbial gene expression, and in microbial metabolism constitute standard read-outs.
| Method | Read-out |
|---|---|
| 16S rDNA amplicon sequencing | Microbiota composition |
| Shotgun metagenomic sequencing | Microbiota composition and predictive functional analysis |
| Metatranscriptome sequencing | Microbial gene expression |
| qPCR | Quantitative detection of specific microorganisms and genes |
| Chromatography (Gas/Liquid) | Analysis of microbial metabolites and biochemical compounds |
Table 1: Overview of the analytical methods that are most commonly applied in i-screen experiments.
The composition of the microbiota is assessed rapidly and efficiently with DNA-based technology. Next generation sequencing techniques, including both marker gene-based sequencing (e.g. 16S rDNA) and the more extensive shotgun metagenomic sequencing, are often used after an i-screen experiment, as they provide in-depth taxonomic characterization of complex microbial communities [8].
Transcription patterns are tracked by studying gene expression through metatranscriptomics or qPCR. Because of its specificity, qPCR allows to track certain genes and bacteria and to answer distinct research questions. For example, species with relevance for human health, such as bifidobacteria, lactobacilli, enteropathogenic E.coli, and toxigenic C. difficile and C. perfringens, can be specifically targeted and studied [9]. qPCR can also be used to target antibiotic resistance and virulence genes, for example to evaluate the effects of antimicrobial strategies on the occurrence and development of antibiotic resistance.
Microbial metabolism is studied by analyzing samples with gas chromatography mass spectrometry (GC-MS) or liquid chromatography mass spectrometry (LC-MS) [9]. Key products of microbial metabolism that are routinely measured with these techniques include short chain fatty acids (SCFAs) and branched chain fatty acids (BCFAs) [8,9], which have been demonstrated to play important roles in the maintenance of health in vivo. Other relevant metabolic pathways include the formation of secondary bile acids and tryptophan metabolism, which is linked to brain function. High-performance Anion Exchange Chromatography (HPAEC) is also very useful for studying microbial degradation of specific molecules, for example the breakdown of prebiotic dietary oligosaccharides or the conversion of drugs to their active forms [8,10-12].
The i-screen can provide a better understanding of the gut microbiota and its functioning, specifically with regard to the interactions with different compounds. This knowledge can inform and guide early research phases in functional food or drug development. Importantly, the i-screen has the potential to reduce the number of test animals, and to provide information that can be used to optimize the human studies needed during the development process. Furthermore, the use of this technology does not require ethical approval. These factors significantly contribute to shorter time to market and considerable cost saving during product development (Table 2).
| Some of the questions that can be addressed using the i-screen |
|---|
| How does a test compound influence the microbiota composition? What are the effects of the compound on the activity of the microbiota? What are the effects of the compound on the microbiota of different individuals/population groups? What is the effective dose of the compound? How do different products and combinations of products differ in their effect on the microbiota? What bacterial species are responsible for specific metabolic effects? What is the mechanism of action of the compound on the microbiota? |
Table 2: Questions that can be addressed using the i-screen.
The i-screen can be applied for several different purposes:
Microbiota s(t)imulation studies
The intestinal microbiota supports several metabolic functions and is involved in many mechanisms supporting the maintenance of health or leading to the development of disease. Using the i-screen one can not only simulate the microbiota, but also stimulate it to create a diseased state. For example, specific pathogenic bacteria can be added to the microbiota in the i-screen, and the following response of gut microorganisms to disease-inducing species can be monitored. This type of experiment can yield new insights in the disease-inducing mechanisms triggered by certain pathogens, and in general it can help elucidate role of the gut microbiota in the maintenance of health.
Pharmaceutical compounds development
The search for novel pharmaceuticals is a complex, expensive and time-consuming process, with extensive clinical tests required to demonstrate the working mechanisms and to investigate the side effects of new drugs. The gut microbiota can influence pharmacokinetic efficacy and toxicity profiles, and in turn some drugs can affect the composition and/or the metabolic activity of the gut microbiota [13]. The i-screen can rapidly identify drugs that are susceptible to transformation by microbial metabolism. Unknown metabolites that are produced as a result of these transformations can also be readily identified, while in traditional clinical studies metabolite profiling is only done in later phases of research, usually phase II and III. By allowing to test a large number of conditions simultaneously while avoiding lengthy regulatory processes, i-screen experiments can render the drug development process more efficient by saving time and money.
Food ingredient research
Novel functional food ingredients also require extensive studies before becoming available on the market. Usually, many substances need to be screened and both in vitro testing and human studies are required to demonstrate their safety and to validate their efficacy. As for pharmaceutical compounds, the i-screen technology can support decision making early in the development stage of food ingredients, thereby shortening the time to market of novel products. The i-screen can also be used to guide the development of products targeted for specific population groups, for example different age classes (from newborn to elderly) and health conditions (such as obesity or inflammatory diseases) that can be easily modeled in this in vitro platform.
In order to standardize the tests and to account for interindividual variability, pools of fecal material are generally used in i-screen experiments, allowing the extrapolation of results to the general population. On the other hand, focus on distinct patient populations is necessary to characterize the gut microbiota and its functionality in specific disease conditions. Since each individual has a unique microbiota, a personalized approach in microbiome studies may be advisable. We anticipate that the i-screen, as well as similar existing platforms, will be increasingly used to investigate differences between the microbiota of different individuals, and differences in responses to specific compounds. In the long term, this type of approach could support the choice for a specific medicine or prebiotic compound on an individual basis, with the ultimate goal to personalize disease treatments and health and nutritional advice [14].
The relationship between the gut microbiota and health constitutes the basis for the development of novel strategies to promote gut health through microbial modulation [15]. For example, while pre- and probiotics traditionally targeted Bifidobacterium and Lactobacillus species, they have now expanded to new taxa such as Clostridium and Akkermansia, offering opportunities for the development of new products. By providing a platform to test the efficacy of compounds and to study their effects on gut microorganisms, the i-screen can direct us towards novel approaches to support health by modulating the microbiota [16].
The i-screen is a powerful and flexible system to study the human gut microbiota in vitro, and can provide useful information on microbial metabolism. However, it is currently limited in its capacity to provide information about the metabolism of the host. In order to better understand the complex host-microbe interactions that characterize the intestinal environment, integration with in vitro host models is desirable. Different research groups are working on developing models to study hostmicrobe interactions and future developments of the i-screen will also be oriented in this direction.
Citation: Schuren F, Agamennone V, Keijser B, Abeln E, der Vossen J, Montijn R (2019) The i-screen: A Versatile Preclinical Platform for Gut Microbiota Studies. J Prob Health. 7:212. DOI: 10.35248. 2329-8901.19.7.212
Received: 25-Aug-2019 Accepted: 09-Sep-2019 Published: 16-Sep-2019 , DOI: 10.35248/2329-8901.19.7.212
Copyright: © 2019 Schuren F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.