Drug Designing: Open Access
Open Access
ISSN: 2169-0138
ISSN: 2169-0138
Research - (2021)Volume 10, Issue 2
Nano-organometallic compounds; Elemental and spectral analyses; Biological activity
Acetaminophen (an international name used in the USA) is derived from its chemical name N-acetyl-para-aminophenol. This drug has a long history and, as it often happens with important discoveries, it was found by chance. Acetaminophen is one of the most popular and most commonly used analgesic and antipyretic drugs around the world, available without a prescription, both in mono- and multi-component preparations [1-5]. It is the drug of choice in patients that cannot be treated with Non-Steroidal Anti-Inflammatory Drugs (NSAID), such as people with bronchial asthma, peptic ulcer disease, hemophilia, salicylate-sensitized people, Children under 12 years of age, pregnant or breastfeeding women [6-8]. It is recommended as a first-line treatment of pain associated with osteoarthritis. The mechanism of action is complex and includes the effects of both the peripheral (COX inhibition), and central (COX, serotonergic descending neuronal pathway, L-arginine/NO pathway, cannabinoid system) antinociception processes and "redox" mechanism. Nanotechnology refers to the creation and utilization of materials whose constituents exist at the nanoscale and by convention, be up to 100 nm in size [9]. Nanotechnology explores electrical, optical, and magnetic activity as well as structural behavior at the molecular and sub-molecular level. It has the potential to revolutionize a series of medical and biotechnology tools and procedures so that they are portable, cheaper, safer, and easier to administer [10]. Due to their exceptional properties including antibacterial activity, high resistance to oxidation and high thermal conductivity, nanoparticles have attracted considerable attention in recent years. Nanoparticles in the form of organometallic compounds can be synthesized chemically or biologically. Viruses are infectious agents that affect cells of almost all types of organisms. They are the smallest and simplest of all the microbes [11-15]. All viruses contain DNA or RNA. This genetic material is enclosed by a protein shell called a capsid. Many viruses also have an outer membrane surrounding them. Most viruses are shaped like rods or spheres. However, some viruses have a very unique appearance. Viruses need a host cell to reproduce. The virus will invade a healthy cell and use it to make more viruses. The host cell is usually killed in this process. Once inside the host cell, the virus injects its DNA (RNA) into the cell. The viral DNA takes control and makes copies of it-self leading to more and more viruses. The new viruses then burst out of the cell and invade others, over and over. Most viruses cannot be treated. A person’s immune system must fight it without the typical help of medicines. However, vaccines (immunizations you can get in order to prevent a virus from invading your cells and making you sick) and some anti-viral drugs may be used to control or prevent the spread of viral diseases. Viral diseases are among the most widespread illnesses in humans [16-18]. These illnesses range from mild fevers to some forms of cancer and include several other severe and fatal diseases. Transmission of these illnesses varies .Some are transmitted by human contact, while others are transmitted through water, an insect bite or are airborne. In this manuscript, new nano-organometallic compounds analogue to acetaminophen drug have been prepared and characterized using elemental and spectroscopic methods. Antimicrobial activity of these compounds has been studied as well as cytotoxic effect on experimental rats [19-21].
Materials
The starting chemicals were of analytical grade and used without further purification.
Physical and spectroscopic techniques
The characterization of the ligand and its compounds carried out using various spectroscopic techniques such as:-Elemental analyses (C, H, N and Cl) were performed by analytical laboratory of Cairo University, Egypt.
Molar conductivity
The molar conductivity of 10-3 M of compounds in dimethylsulfoxide (DMSO) was determined using Bibby conductimeters MCI at room temperature. Molar conductivities were calculated according to the following equation:
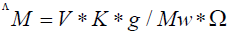
Where: ΛM=molar conductivity (ohm-1cm2mol-1)
V=volume of the solution (100 cm3)
K=cell constant: 0.92 cm-1
Mw=molecular weight of the complex
g=grams of complex dissolved in 100 cm3 solution
Ω=resistance measured in ohms.
Mass spectra
The mass spectra of the compounds were recorded on JEOL JMS-XA- 500 mass spectrometer.
Thermal analyses
DTA and TGA were carried out on a Shimadzu DT-30 thermal analyzer in nitrogen atmosphere, from room temperature to 600°C at a heating rate of 10°C per minute.
1H-NMR spectra
The 1H-NMR spectra were recorded on a JEOL EX-270 MHZ FTNMR spectrometer in deuterated dimethylsulfoxide (DMSO-d6) as a solvent. The chemical shifts were measured relative to the solvent peaks.
IR spectra
The infrared spectra of the ligand and its compounds were recorded on Perkin Elmer's infrared spectrometer 681 using KBr or CsBr discs.
Electronic absorption spectra
The electronic absorption spectra of the compounds were recorded on UNICO SQ-4802 UV/Vis. Double beam spectrophotometer (190-1100 nm) using 1 cm quartz cell using DMSO as a solvent.
Magnetic susceptibility
The magnetic susceptibilities of the solid compounds in the solid state were measured in a borosilicate tube with a Johnson Matthey [8] at room temperature using the following equations:
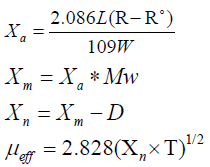
Where
Χa=mass susceptibility; L=sample length in cm; R=tube+sample reading; R°=empty the reading; W=mass of the sample; Χm=molar susceptibility; Mw=molecular weight; Χn=corrected molar susceptibility; D=diamagnetic corrections; μeff=effective magnetic moment; T=room temperature in Kelvin.
Determination of metal content
Metal content was determined using colorimetric method on HACH DR 5000 spectrophotometer [22-24].
ESR spectra
The solid ESR spectra of some compounds were recorded with ELEXSYS E500 Bruker spectrometer in 3 nm Pyrex Tubes at 25°C. Diphenylpicrl-hydrazide (DPPH) free radical was used as a g-marker for the calibration of the spectra. The equation used to determine g-values was g=(gDPPH) (HDPPH)/H Where: gDPPH=2.0036; HDPPH=magnetic field of DPPH in gauss; H=magnetic field of the sample in gauss.
Preparation of the ligand (1)
It was prepared by refluxing with stirring for two hours of p-hydroxymethyl benzoate (10.0 g, 1.914 mol) with o-phenylene diamine (6.2 g, 1.913 mol) in 30 cm3 ethanol. The solid product which formed left to cool at room temperature, filtered off, washed with ethanol and dried in air. The solid hydrized p-hydroxy-benzoic-1,2- phenylene diamine (15.0 g, 0.188 mol) dissolved in 35 cm3 ethanol, acetic anhydride (6.7 g, 0.187 mol) was added. Reflux the mixture with stirring for another two hours and then left the product to cool at room temperature, filtered off the formed precipitate and leave it to dry at room temperature. Recrystallize the product in ethanol and dried to give the final organic ligand.
Preparation of ligand (1) is shown in Figure 1.
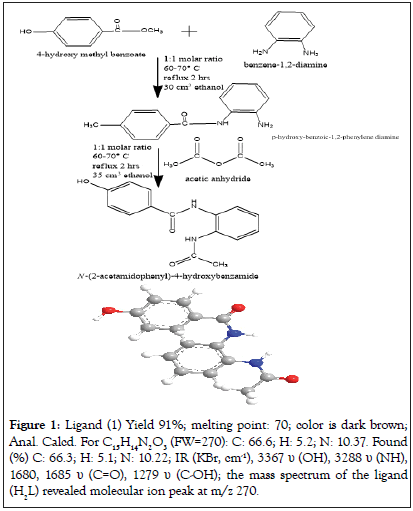
Figure 1: Ligand (1) Yield 91%; melting point: 70; color is dark brown; Anal. Calcd. For C15H14N2O3 (FW=270): C: 66.6; H: 5.2; N: 10.37. Found (%) C: 66.3; H: 5.1; N: 10.22; IR (KBr, cm-1), 3367 υ (OH), 3288 υ (NH), 1680, 1685 υ (C=O), 1279 υ (C-OH); the mass spectrum of the ligand (H2L) revealed molecular ion peak at m/z 270.
The mass spectrum of the ligand (H2L) revealed molecular ion peak at m/z 270. Analytical data for ligand (1) and its compounds are given in Table 1.
| S. No. | Ligand/compounds | Color | FW | M.P (°C) | Yield (%) | Anal./Found (Calc.) (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| C | H | N | M | Conductivity Λ | ||||||
| 1 | [HL] C15H14N2O3 | Dark brown | 270 | 70 | 91 | 66.3 (66.6) | 5.1 (5.2) | 10.22 (10.37) | - | - |
| 2 | [(HL) Cu (OAc)2(H2O) ].2H2O C19H26CuN2O10 | Black | 505.5 | 88 | 92 | 45.22 (45.1) | 5.13 (5.1) | 5.55 (5.5) | 12.75 (12.5) | 4.3 |
| 3 | [(HL) Cd (OAc)2(H2O) ].2H2O C19H26CdN2O10 | Brown | 554.5 | 84 | 70 | 41.35 (41.1) | 4.7 (4.6) | 5.01 (5.05) | 20.33 (20.2) | 5 |
| 4 | [(HL) Ni (OAc)2(H2O) ].2H2O C19H26NiN2O10 | Light green | 507.39 | 93 | 85 | 44.66 (44.9) | 5.13 (5.1) | 5.53 (5.5) | 11.67 (11.55) | 4.1 |
| 5 | [(HL) Cu (SO4) (H2O)2 ].2H2O C15H22CuN2O11S | Black | 501.5 | 92 | 89 | 35.87 (35.9) | 4.47 (4.4) | 5.62 (5.6) | 12.92 (12.7) | 7 |
| 6 | [(HL) Co (SO4) (H2O)2 ].2H2O C15H22CoN2O11S | Black | 496.9 | 95 | 79 | 36.29 (36.2) | 4.53 (4.4) | 5.49 (5.6) | 11.97 (11.6) | 6.5 |
| 7 | [(HL) Zn(NO3)2 (H2O)]. H2O C15H18ZnN4O11 | orange | 495.3 | 80 | 68 | 36.99 (36.3) | 3.69 (3.6) | 11.66 (11.3) | 13.39 (13.2) | 4.8 |
Note: *m-1cm2mol-1
Table 1: Analytical and physical data of the ligand 1 and nano-metal compounds.
Preparation of organometallic compounds (2)-(7)
The metal compounds (2)-(7) were prepared by refluxing with stirring a suitable amount of a hot ethanolic solution (30 cm3) of the following metal salts: Cu(OAc)2.H2O (0.29 g, 0.053 mol) compound (2), Cd(OAc)2.4H2O (0.349 g, 0.054 mol) compound (3), Ni(OAc)2.4H2O (0.399 g, 0.0534 mol) compound (4), CuSO4.5H2O (0.358 g, 0.0534 mol) compound (5), CoSO4.7H2O (0.450 g, 0.0534 mol) compound (6), Zn(NO3)2.4H2O (0.304 g, 0.0533 mol) compound (7), with a hot ethanolic solution of the ligand (1) (0.5 g, 0.0534 mol) in 30 cm3 ethanol for 1-3 hours range depending on the anion used. The precipitates which formed were filtered off, washed with ethanol then by diethyl ether and dried in vacuum desiccators over P4O10. The analytical and physical data are shown in Table 1.
Biological activity
Antimicrobial activity: Antimicrobial activity of the tested compounds was assessed against gram positive bacteria (Staphylococcus aureus), gram negative bacteria (Escherichia coli) and fungi (Aspergillus flavus) and (Candida albicans) using disc diffusion method [25]. Standard discs of Ampicillin (Antibacterial agent), Amphotericin B (Antifungal agent) served as positive controls for antimicrobial activity. The test compounds were dissolved in DMSO to give concentrations 250, 200, 175, 150 and 125 ppm and a DMSO poured disc was used as negative control. The bacteria were subcultured in nutrient agar medium, which was prepared using peptone, beef extract, NaCl, agar and distilled water. The Petri dishes were incubated for 48 hours at 37°C. The standard antibacterial drug tetracycline was also screened under similar conditions for comparison. The zone of inhibition was measured in millimeters carefully. All determination was made in duplicate for each of the compounds. An average of the two independent readings for each compound was recorded.
Antivirsus activity: Nano-Cd(II) compound (3) has been prepared in concentration 1 × 10-5ml mol/L (DMF/Saline). Compound spray is used for two successive days with good coverage of infected leaf surface including young, recently unfolded leaves. After one week, we photographed the infected plant without treatment with the compound and plant that was sprayed with the compound.
The ligand (1) and its metal compounds (2)-(7) are stable at room temperature, non-hydroscopic and are insoluble in common solvents such as ethanol, acetone, water and chloroform but soluble in DMF and DMSO. The compounds are in nano-form and stable up to 50°C. The elemental analysis confirmed that, all compounds are composed of the ligand and metal ions with molar ratio equal to 1L:1M. Many attempts were made to grow a single crystal, but unfortunately, they failed until now. Organo-metallic compounds are found in nano form having size range from (5.68-9.47) make different properties of these compounds. The analytical, physical and spectral data are presented in Tables 1-6 which are compatible with the suggested structures as shown in Figure 2. Electron microscopic image of compound (3) is showed in Figure 3.
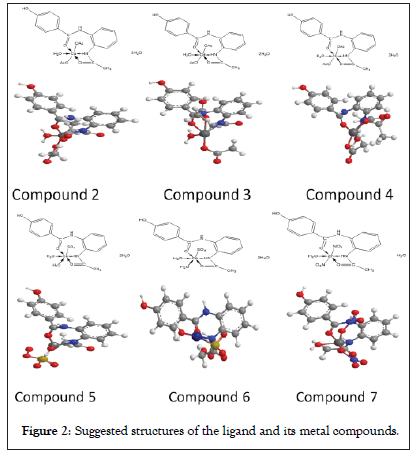
Figure 2: Suggested structures of the ligand and its metal compounds.
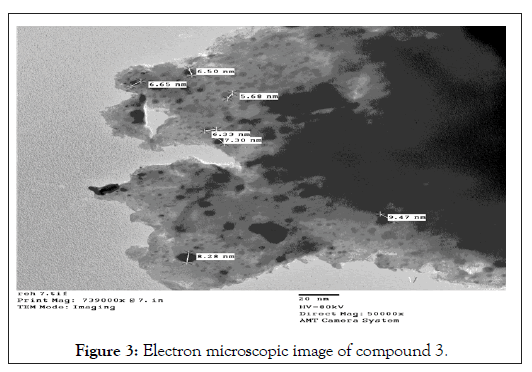
Figure 3: Electron microscopic image of compound 3.
The molar conductivity
The molar conductivities of 1 × 10-3 M solutions of the compounds in DMSO at room temperature were found in the range 4.3-7.0 ohm-1 cm2 mol-1 indicating the non-electrolytic in nature of all compounds. These low values commensurate the coordination of the anions to the metal ions [24-26].
Mass Spectra of the Ligand [H2L], (1) and some of its nano-metal compounds
The mass spectrum of ligand [H2L], (Table 2a) showed the molecular ion peak at m/z 270 amu, confirming its formula weight (F.W. 270).The mass fragmentation patterns observed at m/z=52, 65, 77, 93, 121, 132, 200 and 270 amu correspond to C3H2N, C4H3N, C5H3N, C5H3NO, C6H3NO2, C6H14NO2, C9H16N2O3 and C15H14N2O3 moieties, respectively, supported the suggested structure of the ligand.
| Ligand | m/z | Rel. Int. | Fragment |
|---|---|---|---|
| Ligand 1 | 52 | 20 | C3H2N |
| 65 | 55 | C4H3N | |
| 77 | 18 | C5H3N | |
| 93 | 15 | C5H3NO | |
| 121 | 76 | C6H3NO2 | |
| 132 | 100 | C6H14NO2 | |
| 200 | 18 | C9H16N2O3 | |
| 270 | 15 | C15H14N2O3 |
Table 2a: Mass spectrum of the ligand 1.
The mass spectrum of compound (2) (Table 2b) showed the molecular ion peak at m/z 505 amu, confirming its formula weight (F.W. 505).The mass fragmentation patterns observed at m/z=53, 60, 80, 93, 108, 121, 152, 192, 310, 326, 368, 438 and 505 amu correspond to C3HO, C3H8O, C3H12O2, C4H13O2, C4H14NO2, C5H15NO2, C5H16N2O3, C8H20N2O3, C11H22N2O4Cu, C11H22N2O5Cu, C13H24N2O6Cu, C16H26N2O8Cu and C19H26N2O10Cu moieties, respectively, supported the suggested structure of the compound.
| Compound | m/z | Rel. Int. | Fragment |
|---|---|---|---|
| Compound 2 | 53 | 18 | C3HO |
| 60 | 96 | C3H8O | |
| 80 | 27 | C3H12O2 | |
| 93 | 20 | C4H13O2 | |
| 108 | 51 | C4H14NO2 | |
| 121 | 100 | C5H15NO2 | |
| 152 | 39 | C5H16N2O3 | |
| 192 | 27 | C8H20N2O3 | |
| 310 | 25 | C11H22N2O4Cu | |
| 326 | 34 | C11H22N2O5Cu | |
| 368 | 38 | C13H24N2O6Cu | |
| 438 | 26 | C16H26N2O8Cu | |
| 505 | 25 | C19H26N2O10Cu |
Table 2b: Mass spectrum of the compound 2.
However, the mass spectrum of the compound (7) (Table 2c) showed the molecular ion peak at m/z 495 amu, confirming its formula weight (F.W. 495).The mass fragmentation patterns observed at m/z=54, 66, 80, 92, 108, 121, 133, 150, 163, 192, 236, 279, 300, 319, 402, 456, 480 and 495 amu correspond to C3H2O, C4H2O , C4H2NO, C5H2NO, C5H2NO2, C6H3NO2, C7H3NO3, C7H4NO3, C8H5NO3, C8H6N3O3, C9H6N3O5, C10H6N3O7, C10H11N3O8, C10H14N3O9, C10H16N3O10Zn, C12H16N4O11Zn, C14H16N4O11Zn, and C15H18N4O11Zn moieties, respectively, supported the suggested structure of the compound.
| Compound | m/z | Rel. Int. | Fragment |
|---|---|---|---|
| Compound 7 | 54 | 19 | C3H2O |
| 66 | 24 | C4H2O | |
| 80 | 43 | C4H2NO | |
| 92 | 22 | C5H2NO | |
| 108 | 100 | C5H2NO2 | |
| 121 | 20 | C6H3NO2 | |
| 133 | 33 | C7H3NO3 | |
| 150 | 20 | C7H4NO3 | |
| 163 | 23 | C8H5NO3 | |
| 192 | 18 | C8H6N3O3 | |
| 236 | 22 | C9H6N3O5 | |
| 279 | 15 | C10H6N3O7 | |
| 300 | 15 | C10H11N3O8 | |
| 319 | 12 | C10H14N3O9 | |
| 402 | 18 | C10H16N3O10Zn | |
| 456 | 16 | C12H16N4O11Zn | |
| 480 | 15 | C14H16N4O11Zn | |
| 495 | 12 | C15H18N4O11Zn |
Table 2c: Mass spectrum of the compound 7.
IR spectra
The IR spectra of the ligand (1) showed a strong vibration band located at 1685 cm-1 which assigned to the carbonyl ν(C=O) of the acetyl group , whereas the weak and broad bands which observed at 3467 and 3288 cm-1 are assigned to the stretching vibrations of the aromatic hydroxyl and ν(NH) groups. The broad bands appeared at 3560-3170 and 3160-2600 cm-1 ranges are assigned to inter- and intramolecular hydrogen bondings [27-31]. These observations confirmed the presence of the ligand in the ketonic form in the solid state [29-34]. The spectrum of the ligand also showed band at 1625 cm-1which may be assigned to keto of amide ν(C=O) groups [32]. By comparing the IR spectral data of the nano-compounds with that of the free ligand. It is noted that, in all compounds the appearance of a new band in the range of 1678-1670 cm-1 which may be assigned to the C=O acetyl group and this band shifted to lower value confirming coordination of this group to the metal ion [31]. In the spectra of all compounds, the absorption frequency of amide carbonyl group ν(C=O) was shifted to lower frequency side and appeared in the region 1614-1607 cm-1 range with lowering its intensity confirmed the coordination of oxygen atom of amide ν(C=O) with the metal ions without undergoing enolization. Also, it was observed that, the presence of absorption band due to hydroxyl group which appeared in the spectra of compounds at 3465-3450 cm-1 range indicated that, this group is not coordinated to the metal ion Table 3 [35]. The appearance of new bands appeared in the ranges 626-605, and 565-505 cm-1 ranges for different compounds may be assigned to the ν(M-O), and ν(M←N), respectively [30-37]. These results confirmed that, the bonding of the ligand with the metal ions occurred via carbonyl oxygen of acetyl and amide group and NH nitrogen atom [38]. In the case of acetate compounds (2)-(4) there are two new bands appeared in the 1443-1440 and 1315-1314 cm-1 ranges which are attributed to the symmetric and asymmetric stretching vibration of the acetate group. The separation values (Δ) between νasym(COO-) and νsym.(COO-) in these compounds suggesting the coordination of acetate group in as a monodentate fashion [39,40]. In the case of sulfate compounds (5) and (6), there are new bands appeared in the 1234-1231, 1164-1100 and 697-688 ranges for the complexes, respectively. These bands indicate that, the sulfate ion is coordinated to the metal ion. Compound (7) showed bands at 1373, 1235, 800 and 697 cm-1 indicating coordination of nitrate group to the metal ion in unidentate fashion [38-42]. The above results together with elemental analyses indicate that, the ligand behaved as neutral tridentate fashion bonded to the metal ions through carbonyl oxygen atoms of acetyl and amide and NH groups [39-47].
| S. No. | ν(H2O) | ν(OH) | υ(H-bonding) | ν(C=O) Acetyl | ν(C-OH) | ν(C=O) Amide | ν(NH) | ν(Ar) | ν(OAc)/SO4/NO3) | υ(M-O) | υ(M-N) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | - | 3467 | (3560-3170), (3160-2600) | 1685 | 1279 | 1625 | 3288 | (1550, 1511), (850, 748) | - | - | - |
| 2 | (3470-3270), (3260-3100) | 3460 | (3560-3210), (3200-2750) | 1670 | 1279 | 1607 | 3273 | (1540, 1520), (851, 769) | 1442, 1315 | 626 | 560 |
| 3 | (3460-3290), (3240-3110) | 3450 | (3540-3200), (3180-2740) | 1671 | 1280 | 1610 | 3270 | (1538, 1518), (852, 768) | 1443-1315 | 618 | 561 |
| 4 | (3450-3280), (3240-3105) | 3455 | (3550-3220), (3190-2760) | 1671 | 1279 | 1606 | 3265 | (1536, 1517), (852, 767) | 1440-1314 | 620 | 556 |
| 5 | (3528-3313), (3300-3105) | 3450 | (3573-3200), (3190-2777) | 1678 | 1280 | 1614 | 3290 | (1532, 1514), (851, 769) | (1231,1159),(1110,697) | 613 | 562 |
| 6 | (3520-3320), (3310-3115) | 3465 | (3571-3205), (3195-2782) | 1674 | 1279 | 1612 | 3284 | (1533, 1516), (849, 765) | (1234,1164),(1100,688) | 610 | 561 |
| 7 | (3500-3325), (3315-3175) | 3460 | (3505-3190), (3180-2705) | 1674 | 1277 | 1614 | 3255 | (1531,1518), (852,769) | (1373,1235),(800,697) | 611 | 561 |
Table 3: IR frequencies of the bands (cm-1) of the ligand 1 and its nano-metal compounds.
Magnetic moments
The magnetic moments of the nano-metal compounds (2)-(7) at room temperatures are shown in Table 4. Cu(II) compounds (2) and (5) show values at 1.71 and 1.69 B.M corresponding to one unpaired electron in the a d(x2-y2) ground state in an octahedral structure. The low values than 1.73 B.M is due to spin – spin interactions takeplace between Cu(II) ions [43-48]. Cd(II) complex (3) Zn(II) compound (7) showed diamagnetic property [53-56]. Ni(II) compound (4) showed value equal to 2.93 B.M. The lowering in the value may be assigned to the interaction between the two nickel atoms via hydrogen bondings or molecular association [43- 49]. The magnetic moment of Co(II) compound (6) equal to 4.75 B.M, indicating octahedral structure [50-52].
| S. No. | λmax (nm) | eff in B.M. | ν2/ν1 |
|---|---|---|---|
| 1 | 295 nm (log e =3.98), 315 nm ( log e =4.25) 335(log e =4.59) | - | - |
| 2 | 280, 305, 315, 450, 590, 000 | 1.71 | - |
| 3 | 290, 310, 320 | Diamagnetic | - |
| 4 | 288, 310, 320, 462, 570, 610, 740 | 2.93 | 1.56 |
| 5 | 287, 302, 318, 475, 575, 608 | 1.69 | - |
| 6 | 292, 305, 315, 435, 562, 610 | 4.75 | - |
| 7 | 298, 305, 325 | Diamagnetic | - |
Table 4: The electronic absorption spectral bands (nm) and magnetic moments (B.M.) for the ligand 1 and its nano-compounds.
Electronic spectra
The electronic spectral data for the ligand (1) and their metal compounds in DMSO are summarized in Table 4. The electronic absorption spectra of the ligand showed three bands located at 295, 315 and 335 nm respectively. The first one assigned to intraligand π → π*transition in the hydroxyl moiety which is nearly unchanged on complexation. The second and third bands may be assigned to n → π* and charge transfer transitions of the NH and carbonyl groups. The location of these bands was found to be shifted to higher energy on complexation. This finding indicated to the participation of these groups in bonding and coordination with metal ions [28,46]. Copper(II) compounds (2), and (5) showed bands in the 318-280, 475-450, 590-575 and 610-608 nm ranges, were assigned to intraligand transition O → Cu, charge transfer,2B1 → 2E and 2B1 →2B2 transitions, indicating a distorted octahedral structure [57-59]. The electronic spectrum nickel(II) compound (4) showed bands at 288, 310, 320, 462, 570, 610 and 740 nm, which are assignable to intraligand transition 3A2g (F) → 3T1g (p) (3), 3A2g (F) → 3T1g (F) (2) and 3A2g (F) → 3T1g (F) (1) transitions respectively, assuming the octahedral Ni(II) compound [54,55]. The (ν2/ν1) ratio for the compound was less than the usual value 1.6 indicating distorted octahedral Ni(II) compound, the (ν2/ν1) value equal to 1.56, indicating distorted octahedral structure. The electronic spectrum of Co(II) compound(6) showed bands at 292, 305, 315, 435, 562 and 610 nm. The first three bands are within the ligand and indicating intraligand transition. However, the other bands are due to 4T1g (F) →4T1g (p), 4T1g (F) →4A2g (F), 4T1g (F) →4T2g(F), transition respectively, indicating distorted octahedral structure around the Co(II) ion [60]. Cd(II) compound (3) and Zn(II) compound (7) showed bands were due to intraligand transitions.
Electron Spin Resonance (ESR)
The ESR spectral data for copper (II) compounds (2) and (5) were presented in Table 5. The spectra of copper(II) compounds were characteristic of species d9 , configuration having axial type of a d(x2-y2) ground state which is the most common for copper(II) compounds. The compounds showed g||>g┴>2.0023, indicating octahedral geometry around copper(II) ion. The g-values are related by the expression G=(g||-2)/ (g┴ -2), where (G) exchange coupling interaction parameter (G). If G<4.0, a significant exchange coupling is present, whereas if G value >4.0, local tetragonal axes are aligned parallel or only slightly misaligned. Copper(II) compounds showed 3.00 and 2.70 values indicating spin-exchange interactions takeplace between copper(II) ions. This phenomenon is further confirmed by the magnetic moments values Table 4. The g||/A|| value is also considered as a diagnostic term for stereochemistry, the g||/A|| values are 155.7 and 182.5 which are expected for distorted octahedral compounds [60-64]. The g-values of the copper(II) compounds with a 2B1g ground state (g||>g┴) may be expressed by

Where k|| and k┴ are the parallel and perpendicular components respectively of the orbital reduction factor (K), is the spin-orbit coupling constant for the free copper, ΔExy and ΔExz are the electron transition energies of 2B1g→2B2g and 2B1g→2Eg. From the above relations, the orbital reduction factors (K||, K┴, K), which are measure terms for covalency, can be calculated. For an ionic environment, K=1; while for a covalent environment, K<1. The lower the value of K, the greater is the covalency.

K values for the copper(II) compounds are indicating for a covalent bond character. Kivelson and Neiman noted that, for ionic environment g||≥ 2.3 and for a covalent environment g||<2.3. Theoretical work by Smith seems to confirm this view. The g-values reported here showed considerable covalent bond character. Also, the in-plane σ-covalency parameter, α2(Cu) was calculated by
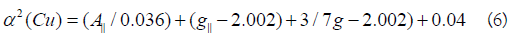
The calculated values suggest a covalent bonding [38,56]. The inplane and out of- plane-bonding coefficients β12 and β22 respectively, are dependent upon the values of ΔExy and ΔExz in the following equations [58].
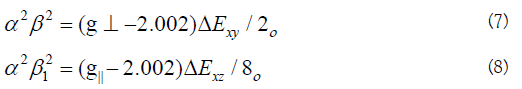
In this work, the compounds showed β12 values indicating a moderate degree of covalency in the in-plane -bonding [56,59]. However, the β2 value for compounds indicating ionic character of the out-of-plane [60-67]. It is possible to calculate approximate orbital populations ford orbitals by
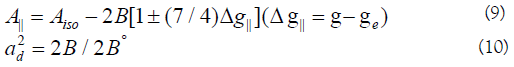
Where A° and 2B° is the calculated dipolar coupling for unit occupancy of d orbital respectively. When the data are analyzed, the components of the [60] Cu hyperfine coupling were considered with all the sign combinations. The only physically meaningful results are found when A⊥ and A|| were negative. The resulting isotropic coupling constant was negative and the parallel component of the dipolar coupling 2B is negative Table 5. These results can only occur for an orbital involving the d(x2-y2) atomic orbital on copper [68-71]. The value for 2B is quite normal for copper(II) compounds. The Aiso value was relatively small. The 2B value divided by 2B° (The calculated dipolar coupling for unit occupancy of d(x2-y2) (235.11 G), using equation (10)) suggests all orbital population are 60, 66, and 57.2% d-orbital spin density, clearly the orbital of the unpaired electron is d(x2-y2). However, Co(II) compound (6) showed isotropic spectrum with 2.12 with covalent bond character [71].
| Complex | g|| | g⊥ | agiso | A || (G) | A⊥ (G) | bAiso (G) | cG | ∆Exy (cm-1) | ∆Exz (cm-1) | K2 ⊥ | K||2 | K2 | K | g||/A|| (cm-1) | α2 | β2 | β2 1 | -2β | a2d (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2.18 | 2.06 | 2.13 | 135 | 7.5 | 50 | 3.00 | 17241 | 21052 | 0.73 | 0.46 | 1.01 | 0.8 | 155.7 | 0.63 | 1.16 | 0.73 | 141 | 60 |
| 5 | 2.19 | 2.07 | 2.11 | 120 | 15 | 30 | 2.7 | 17391 | 21739 | 0.89 | 0.49 | 0.76 | 0.87 | 182.5 | 0.59 | 1.51 | 0.83 | 134 | 57.2 |
| 6 | - | - | 2.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
Table 5: The ESR data for metal (II) compounds.
Thermal analyses (DTA and TGA)
The thermal data of compounds (2), (4), (6) and (7) are listed in Table 6. These compounds were introduced as representative examples. Thermogram of compound (2) showed decomposition in six steps, the first step involving breaking of H-bondings accompanied with endothermic peak at 55°C. In the second step, two molecules of hydrated water molecules were lost endothermically with peak at 80°C accompanied by 6.1% (Calc 5.58%) weight loss. Loss of one coordinated water molecule was recorded in the third step as an endothermic peak at 145°C with 4.21 (Calc. 3.94) weight loss. The 26.52% weight loss (Calc. 26.91%) accompanied by an endothermic peak at 265°C-290°C range was assigned to loss of coordinated 2(OAc) group, whereas the endothermic peak observed at 330°C refers to the melting point of the compound. The final step observed as exothermic peaks at 445°C, 492°C, 550°C and 610°C with 23.0% weight loss (Calc. 22.15%), refers to complete decomposition of the compound which exposed up with the formation of CuO [72-78]. The first step observed in the thermogram of compound (4) involves breaking of H-bondings accompanied with endothermic peak at 58°C. In the second step, two molecules of hydrated water were lost endothermically with peak at 65 accompanied by 6.96% (Calc 7.40%) weight loss. In the third step, two molecules of coordinated water molecules were lost endothermically with a peak at 160°C accompanied by 7.10% (Calc. 7.34%) weight loss. The forth step involved loss of coordinated 2(OAc) group accompanied with an endothermic peak at 285°C-300°C range with 18.63% (Calc. 18.52%). while the endothermic peak appeared at 330°C refers to the melting point of the compound. The final step observed at 470°C, 515°C and 580°C with 26.81% weight loss (Calc. 26.50%) as exothermic peaks, refers to complete oxidative decomposition of the compound which ended up with the formation of NiO. The first step observed in the thermogram of compound (6) involving breaking of H-bondings accompanied with endothermic peak at 48°C. In the second step, two molecules of hydrated water were lost endothermically with peak at 80°C accompanied by 4.1% (Calc 4.31%) weight loss. Loss of one coordinated water molecule was recorded in the third step as an endothermic peak at 120°C with 5.62 (Calc. 5.21%) weight losses. The 31.22% weight loss (Calc. 30.9%) accompanied by an endothermic peak appears at 265°C-282°C was assigned to loss of coordinated SO4 group whereas the endothermic peak observed at 330°C refers to the melting point of the compound. The final step observed at 425°C, 480°C, 530°C and 600°C with 31.4% weight loss (Calc 30.7%), refers to complete oxidative decomposition of the compound which exposed up with the formation of CoO. The first step observed in the thermogram of compound (7) involving breaking of H-bondings accompanied with endothermic peak at 55°C. In the second step, one molecule of hydrated water was lost endothermically with peak at 80°C accompanied by 6.1% (Calc. 5.58%) weight loss. The third step involved loss of one coordinated water molecule accompanied with an endothermic peak at 145°C with 4.21 (Calc. 3.94%). The 26.52% weight loss (Calc 26.91%) accompanied by an endothermic peak appears at 265°C-290°C range was assigned to loss of coordinated 2(NO3) group, whereas the endothermic peak observed at 330°C refers to the melting point of the compound. The final step observed at 445°C, 492°C, 550°C and 610°C with 19.2% weight loss (Calc. 18.50%), refers to complete oxidative decomposition of the compound which exposed up with the formation of ZnO.
| Compound no. molecular formula | Temp. (°C) | DTA (peak) TGA (Wt.loss %) | Assignments | |||
|---|---|---|---|---|---|---|
| Endo | Exo | Calc | Found | |||
| Compound 2 | 55 | endo | - | - | - | Broken of H-bondings |
| 80 | endo | - | 5.58 | 6.1 | Loss of (2H2O) hydrated water molecules | |
| 145 | endo | - | 3.94 | 4.21 | Loss of (H2O) coordinated water molecule | |
| 265,290 | endo | - | 26.91 | 26.52 | Loss of coordinated 2(OAc) group | |
| 330 | endo | - | - | - | Melting point | |
| 445,492,550,610 | - | Exo | 22.15 | 23 | Decomposition process with the formation of (CuO) | |
| Compound 3 | 58 | endo | - | - | - | Broken of H-bondings |
| 65 | endo | - | 7.4 | 6.96 | Loss of (2H2O) hydrated water molecule | |
| 160 | endo | - | 7.34 | 7.1 | Loss of (2H2O) coordinated water molecules | |
| 285,300 | endo | - | 18.52 | 18.63 | Loss of coordinated 2(OAc) group | |
| 330 | endo | - | - | - | Melting point | |
| 470,515,580 | - | Exo | 26.5 | 26.81 | Decomposition process with the formation of (NiO) | |
| Compound 6 | 52 | endo | - | - | - | Broken of H-bondings |
| 80 | endo | - | 4.31 | 4.1 | Loss of (2H2O) hydrated water molecule | |
| 120 | endo | - | 5.21 | 5.62 | Loss of (H2O) coordinated water molecule | |
| 265,282 | endo | 30.9 | 31.22 | Loss of coordinated SO4 group | ||
| 330 | endo | - | - | - | Melting point | |
| 425,480,530,600 | - | Exo | 30.7 | 31.4 | Decomposition process with the formation of (CoO) | |
| Compound 7 | 55 | endo | - | - | - | Broken of H-bondings |
| 80 | endo | - | 5.58 | 6.1 | Loss of (H2O) hydrated water molecule | |
| 145 | endo | - | 3.94 | 4.21 | Loss of (H2O) coordinated water molecule | |
| 265,290 | endo | - | 26.91 | 26.52 | Loss of coordinated 2(NO3) group | |
| 330 | endo | - | - | - | Melting point | |
| 445,492,550,610 | - | Exo | 18.5 | 19.2 | Decomposition process with the formation of (ZnO) | |
Table 6: Thermal analyses for some metal (II) compounds.
Antimicrobial activity
In vitro biological screening tests of the ligand (1) and its metal compounds (2), (3), (4), (5), (6) and (7) carried out as antibacterial and antifungal activity (Figures 4-8). The antibacterial activity was tested against two bacterial strains; Gram-positive Staphylococcus aureus as well as Gram-negative Escherichia coli strains [79,80]. The results compared with standard drugs (Ampicllin (Gram positive) and (Gram negative) as a standard drug. The compounds were also subjected to antifungal activity against Aspergillus flavus and Candida albicans using Amphotericin B as a standard drug [81-83]. The data indicated that, using low concentration against microbes, acetaminophen and ligand no effect was observed. However, the compounds show moderate to high effect, the order for Gram positive is (3)>(6)>standard>(4)>(5)>(2)>(7)>acetaminophen=lig and and Gram negative was (3)> standard>(6)>(4)>(5)>(2)>(7)>a cetaminophen=ligand and Aspergillus flavus was (3)>standard> ac etaminophen=ligand=(2)=(4)=(5)=(6)=(7) and Candida albicans is (3)>standard>(4)>(6)>(5)>(2)> acetaminophen=ligand=(7). In case of high concentation, the order of activity for Gram positive was (3)>standard>ligand>acetaminophen and for Gram negative (3)>standard>ligand>acetaminophen and for Aspergillus flavus was (3)>standard>ligand=acetaminophen and for Candida albicans was (3)>standard>ligand>acetaminophen. In vivo studies showed no side effect was observed for all organs tested. The major groups of antibiotics that are currently in use have three microbial targets: the cell wall synthesis, translational machinery and DNA replication machinery. Unfortunately, microbial resistance can developed against each of these modes of action. Most of the antibiotic resistance mechanisms are irrelevant for nanoparticles (NPs) because the mode of action of NPs is direct contact with the microbial cell wall without the need to penetrate the cell. This raises the hope that NPs would be less prone to promote resistance in microbes than antibiotics. Most microbes exist in the form of a biofilm which often contains diverse species that interact with each other and their environment. Biofilms are specifically microbial aggregates that rely on a solid surface and extracellular products, such as extracellular polymeric substances (EPSs). Microbes move reversibly onto the surface but the expression of EPSs renders the attachment irreversible. Once the microbes settled, synthesis of the microbial flagellum is inhibited and the microbes multiply rapidly resulting in the development of a mature biofilm. At this stage, the microbes are stuck together, forming a barrier that can resist antibiotics and provide a source of systemic chronic infections, thus, biofilm are a serious health threat. Nano materials are materials that have at least one dimension (1-100 nm) in the nanometer scale. NPs have demonstrated broad-spectrum antimicrobial properties. The antimicrobial mechanism of action of NPs is generally described as adhering to one of three models: oxidative stress induction, metal ion release, or non-oxidative mechanism [84-89]. These three types of mechanisms can occur simultaneously [89-91]. Nano Cd(II) compound (3) showed higher activity compared with the standard and other compounds. It may be Cd(II) compound prompt neutralization of the surface electric charge of the microbial membrane and change its penetrability, ultimately leading to microbial death [92-94]. In vitro antibacterial and antifungal activity of the ligand, acetaminophen and compound(3) and also other compounds are shown in Tables 7 and 8.
| Escherichia coli (G-) | Staphyococcus aureus (G+) | Aspergillus flavus | Candida albicans | |
|---|---|---|---|---|
| Control: (DMSO) | 0 | 0 | 0 | 0 |
| Ampicillin: (Antibacterial agent) | 24 | 21 | 0 | 0 |
| Amphotericin: (Antifungal agent) | 0 | 0 | 17 | 21 |
| Acetaminophen | 10 | 0 | 0 | 0 |
| Ligand 1 | 14 | 14 | 0 | 14 |
| Cd(II) compound 3 | 34 | 30 | 34 | 28 |
Table 7: In vitro antibacterial and antifungal activity of the ligand, acetaminophen and compound 3.
| Escherichia coli (G-) | Staphyococcus aureus (G+) | Aspergillus flavus | Candida albicans | |
|---|---|---|---|---|
| Control: (DMSO) | 0 | 0 | 0 | 0 |
| Ampicillin: (Antibacterial agent) | 24 | 21 | 0 | 0 |
| Amphotericin: (Antifungal agent) | 0 | 0 | 17 | 21 |
| Acetaminophene | 0 | 0 | 0 | 0 |
| Ligand(1) | 0 | 0 | 0 | 0 |
| Cu(II) compound 2 | 14 | 15 | 0 | 10 |
| Cd(II) compound 3 | 34 | 30 | 34 | 28 |
| Ni(II) compound 4 | 16 | 17 | 0 | 20 |
| Cu(II) compound 5 | 15 | 16 | 0 | 11 |
| Co(II) compound 6 | 21 | 23 | 0 | 14 |
| Zn(II) compound 7 | 11 | 11 | 0 | 0 |
Table 8: In vitro antibacterial and antifungal activity of some nano-metal compounds.
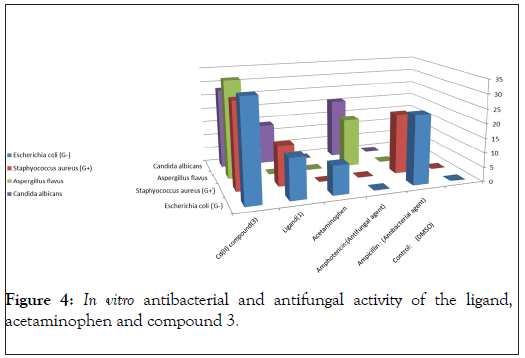
Figure 4: In vitro antibacterial and antifungal activity of the ligand, acetaminophen and compound 3.
Figure 5: In vitro antibacterial and antifungal activity of the ligand and acetaminophen.
Figure 6: In vitro antibacterial and antifungal activity of compound 3.
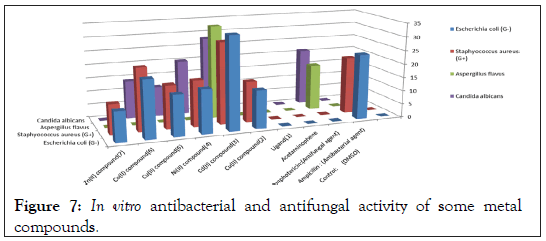
Figure 7: In vitro antibacterial and antifungal activity of some metal compounds.
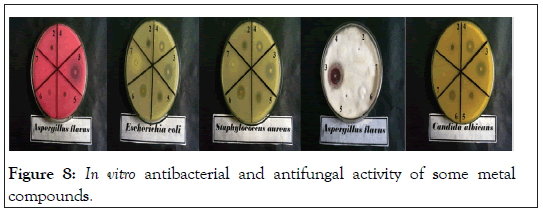
Figure 8: In vitro antibacterial and antifungal activity of some metal compounds.
Transmission electron microscopy characterization (TEM)
TEM images for the colloidal suspensions of Cd(II) compound was obtained. The average diameter of these spherical Cd compound was found to be 7.17 ± 1.30 nm as shown in Figure 9.
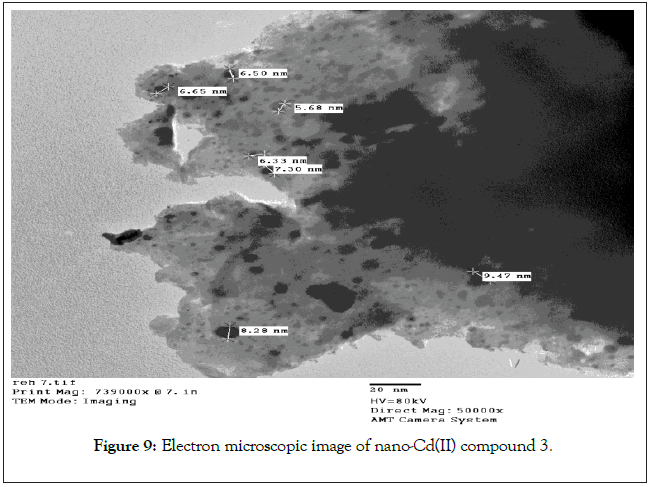
Figure 9: Electron microscopic image of nano-Cd(II) compound 3.
As shown, the compound is present in nano size particles i.e., its particles present in a diameter between 1 nanometer and 100 nanometers in size [95-100]. Nanoparticles can be defined as particles less than 100 nm in diameter that exhibit new or enhanced size-dependent properties compared with larger particles of the same material with many advantages such as:
1. Increased bioavailability
2. Dose proportionality
3. Decreased toxicity
4. Smaller dosage form (i.e., smaller tablet)
5. Stable dosage forms of drugs which are either unstable or have unacceptably low bioavailability in non-nanoparticulate dosage forms.
6. Increased active agent surface area results in a faster dissolution of the active agent in an aqueous environment, such as the human body.
7. Faster dissolution generally equates with greater bioavailability, smaller drug doses, less toxicity.
8. Reduction in fed/fasted variability.
In vivo toxicity studies
Determination of liver functions AST, ALT, and albumin in the serum of normal rats show no significant differences between treated groups and the control group (Tables 9 and 10, Figures 10 and 11).
| Parameters | Control | Cd NPs (1 × 10-5 mmole/L) | Cd NPs (2 × 10-5 mmole/L) |
|---|---|---|---|
| AST (U/l) | 50.20 ± 3.66ac | 48.35 ± 4.34b | 51.36 ± 3.22ac |
| ALT (U/l) | 36.9 ± 4.81a | 27.90 ± 6.91bc | 26.750 ± 2.14bc |
| Alb (g/dl) | 3.90 ± 1.12a | 4.20 ± 1.71bc | 4.170 ± 1.55bc |
| ANOVA: Analysis of Variance; AST: Aspartate aminotransferase; ALT: Alanine aminotransferase; Alb: Albumin; SD: Standard Deviation. Each value is represented as mean ± SD. Data with different superscripts are significantly different at p ≤ 0.05. | |||
Note: aSignificance versus control group; bSignificance versus group treated with Cd NPs with 1 × 10-5 mmole/L; cSignificance versus group treated with Cd NPs with 2 × 10-5 mmole/L.
Table 9: Statistical analysis (ANOVA) for liver function tests in the different groups treated with Cd(II) compound 3 NPs.
| Parameters | Control | Cd NPs (1x10-5 mmole/L) | Cd NPs (2x10-5mmole/L) |
|---|---|---|---|
| B. Urea (mg/dl) | 33.40 ± 2.79a | 36.10 ± 3.55bc | 35.40 ± 2.89bc |
| S. Creatinine (mg/dl) | 0.52 ± 0.11ab | 0.51 ± 0.17ab | 0.565 ± 0.971c |
| ANOVA: Analysis of Variance; SD: Standard Deviation. Each value is represented as mean ± SD. Data with different superscripts are significantly different at p ≤ 0.05. | |||
Note: aSignificance versus control group; bSignificance versus group treated with Cd(II) compound 3 NPs with 1 × 10-5 mmole/L; cSignificance versus group treated with Cd(II) compound 3 NPs with 2 × 10-5 mmole/L
Table 10: Statistical analysis (ANOVA) for renal function tests in the different groups treated with Cd(II) compound 3 NPs.
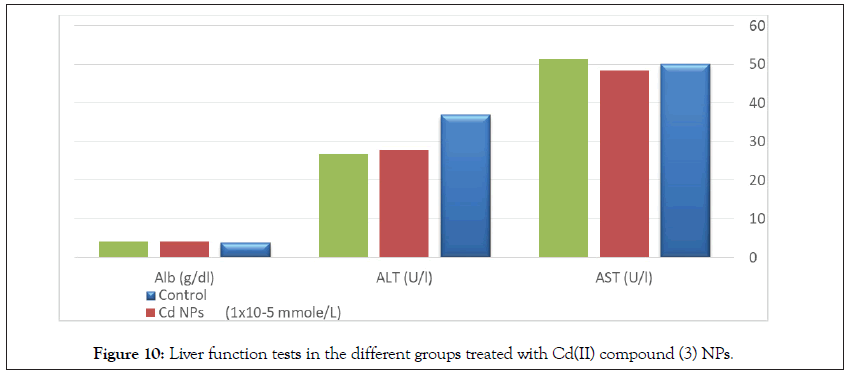
Figure 10: Liver function tests in the different groups treated with Cd(II) compound (3) NPs.
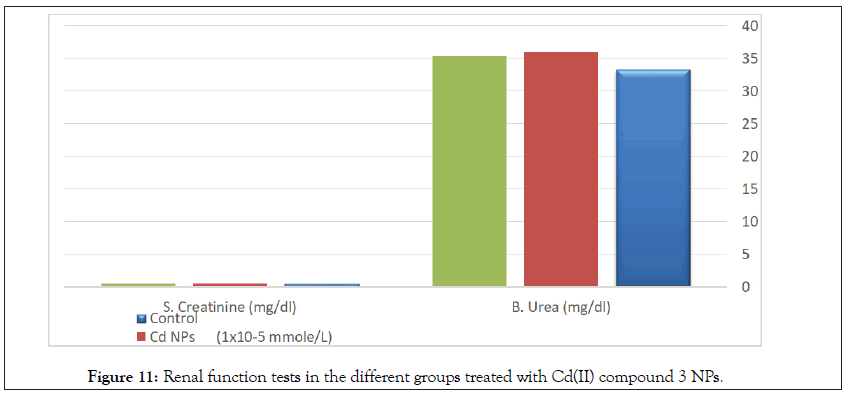
Figure 11: Renal function tests in the different groups treated with Cd(II) compound 3 NPs.
Haematological studies
There are no any significant differences in renal functions or hematological parameters between all tested groups (Table 11, Figure 12) [101].
| Parameters | Control | Cd NPs (6 Weeks) | Cd NPs (12 Weeks) |
|---|---|---|---|
| Hb (g/dl) | 15.70 ± 2.16ab | 15.80 ± 2.89ab | 16.20 ± 1.980c |
| RBCs (× 106/cmm) | 6.11 ± 0.71abc | 6.52 ± 1.12abc | 6.71 ± 1.760abc |
| TLC (× 103/cmm) | 9.90 ± 2.14ac | 10.80 ± 1.77b | 10.10 ± 1.680ac |
| PLTs (× 103/cmm) | 414.42 ± 18.92ac | 448.30 ± 26.41b | 398.81 ± 22.17ac |
ANOVA: Analysis of Variance; Hb: Hemoglobin concentration; RBCs: Red Blood Corpuscles count; T.L.C: Total Leucocytic Count; PLT: Platelets count. Each value is represented as mean ± SD. Data with different superscripts are significantly different at p ≤ 0.05.
Note: aSignificance versus control group; bSignificance versus group treated with Cd(II) compound 3 NPs with 1 × 10-5 mmole/L; cSignificance versus group treated with Cd(II) compound 3 NPs with 2 × 10-5 mmole/L.
Table 11: Statistical analysis (ANOVA) for hematological tests in the different groups treated by Cd(II) compound 3 NPs.
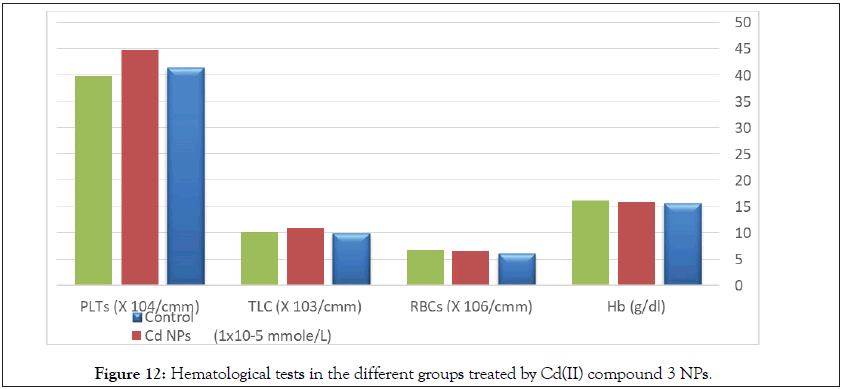
Figure 12: Hematological tests in the different groups treated by Cd(II) compound 3 NPs.
Figure 13 shows the Development stages of medicine.
Figure 13: Development stages of medicine.
Antivirus activity
The great majority has an RNA genome, which is usually small and single strand (SS), but some viruses have double stranded (ds) RNA, SS DNA or ds DNA genomes. Transmission of a virus by insects is a specific biological process. The photographs of the plant before and after treatment are in Figures 14 and 15.
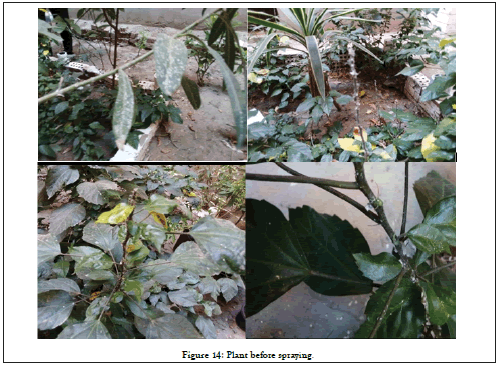
Figure 14: Plant before spraying.

Figure 15: Plant after spraying.
Most plant viruses are rod-shaped with protein discs forming a tube surrounding the viral genome, isometric particles are another common structure. They rarely have an envelope. The great majority have an RNA genome, which is usually small and Single Strand (SS), but some viruses have double stranded (ds) RNA, SS DNA or ds DNA genomes. Transmission of a virus by insects is a specific biological process. The majority of aphids- transmitted viruses affecting plants are transmitted in a non-persistent way. Use Cd(II) nano compound spray to deter aphids from landing and feeding. This method delay virus infection rather than totally prevent it. Spray is very effective in killing aphids breeding on plants and can provide good control of viruses transmitted in a persistent manner. After one week of sprayed the infected plant with Cd(II) nano compound, improvement in leaf surface was observed and growth of the plant was occurred.
Due to the difficulty of testing the nano-Cd(II) compound (3) on animal viruses in our labs, we applied it on plant viruses and impressive results were obtained as shown in plant photographs and this gives us hope for applying it on animal viruses, for example corona virus.
This study was supported by Taif University Researches supporting project number (TURSP-2020/157) Taif University, Taif, Saudi Arabia.
Citation: El-Tabl AS, Abd El-Wahed MM, Ashour AM, Ahmed RAS (2021) The New Acetaminophen Drug in the Form of Nano-Organometallic Compounds as a Bright Future for Antimicrobial Therapy. Drug Des. 10:174.
Received: 28-Dec-2020 Accepted: 11-Jan-2021 Published: 18-Jan-2021 , DOI: 10.35248/2169-0138.21.10.174
Copyright: © 2021 El-Tabl AS, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.