Research - (2022)Volume 6, Issue 5
Background: Palatal rugae are a series of ridges on the hard palate of the mouth with high durability and stability, ensuring their usefulness as a tool for disaster victim identification. Additionally, certain characteristics of palatal rugae are shared by gender or within specific age groups. Currently, forensics odontologists must manually examine palatal rugae imprints to determine an individual’s gender, a time-consuming process vulnerable to bias and human error.
Methodology: This project sought to automate the process of palatoscopy based classification by developing the Rugae Classification Sequence (RCS), a comprehensive tool for gender identification based on features of the palatal rugae. First, data important to palatal rugae–namely rugae length, width, subject age, and gender–were extracted from anonymized images of the palate to provide a bed of training and testing data. The dataset was fed into a series of machine learning algorithms, specifically random decision forest, decision tree classifier, logistic binary classifier, and K-nearest neighbors. Each model then underwent extensive hyperparameter tuning to maximize accuracy and robustness in predicting gender given anatomical properties of the palatal rugae.
Results: On the testing set, the K-nearest neighbor’s algorithm achieved the highest accuracy score and specificity at 65% and 68% respectively, satisfying design requirements of a minimum 60% accuracy in gender classification.
Conclusion: The RCS is a novel application of machine learning in palatoscopy, and with these results, has the potential for large-scale application-indeed, it may provide forensics experts with a more efficient and reliable tool to identify victims given dental remains.
Palatal rugae; Palatoscopy; K-nearest neighbors; Specificity; Forensic odontology; Gender classification
Thesis
The goal of this project is to use machine learning to determine the gender of an individual given the anatomical properties of their palatal rugae. According to the National Census of Medical Examiner and Coroners’ Offices, an estimated 4,400 identified human bodies are received by American medical examiners per year. Over 1,000 of these remains remain unidentified after a single year, hampering national efforts to recover missing persons and productively allocate resources to communities. Moreover, establishing a person’s identity or even determining broad characteristics relating to their gender, age, and race is especially difficult in cases involving terrorist attacks, traffic incidents, and mass disasters. For example, the relatively recent Surfside Condominium collapse, a structural engineering failure outside Miami, Florida, posed an immense challenge to medical examiners-authorities would take an extended 33 days to complete positive IDs for the 98 victims’ remains, most of which were heavily mutilated by crushing debris [1]. Despite the availability of DNA profiling technologies in the modern era, many of these cellular-level techniques remain expensive and time-consuming. Furthermore, dental identification, the process of analyzing teeth to discern an individual’s attributes, can be disrupted through dental treatment prior to the creation of extensive dental records [2]. Consequently, it has become increasingly important that a new method for determining the biological characteristics of a victim, one that is cost-efficient, rapid, and based on durable characteristics of human anatomy, be developed, thus allowing forensic examiners to more effectively limit the population pool for a given human remain.
Forensic odontology involves the management, examination, evaluation, and presentation of dental evidence. Odontologists utilize radiographic examination, comparing antemortem and postmortem pictures to discern the identity of the deceased. Commonly used in situations where physical features are often destroyed or severe trauma is evident, odontologists can compare the combination of decayed, missing, and filled teeth at any fixed point in time to determine the age and identity of the victim. Through dental analysis, odontologists can gain insight into the cause of death, including whether violence was committed against the victim. They can also use sexual dimorphism present in dental development to identify gender–researchers have found significant disparities in canine crown morphology, specifically in the frequency of Distal Accessory Ridges (DAR) on the upper and lower canines, between male and female subjects [3]. However, these modern techniques for dental analysis rely on both the presence of teeth and the assumption that no dental treatment occurred between the creation of antemortem records and time of death.
The palatal rugae are a series of transverse ridges located on the anterior portion of the palatal mucosa, or hard palate. Because they are well protected by the lips, buccal pad of fat, and teeth, these ridges are highly durable, resistant to alterations of structure caused by chemicals, trauma, or disease [4]. Unlike teeth, dental treatments and modifications are not performed specifically on palatal rugae-given the characteristics of a person’s palatal rugae, a forensic examiner can make a more conclusive match to an unknown remain, as the possibility of artificial modifications to the rugae is minimal. The durability and stability of palatal rugae can be attributed to their internal position on the palate, removed from external trauma or growth processes that could alter their shape or structure. Research done at the I.T.S center for dental studies showed that fixed orthodontic treatment did not change palatal rugae patterns, simply modifying the bony structures adjacent to palatal rugae; moreover, correct matches entirely based on palatal rugae pattern occurred for 90.2% of all subjects in the sample [5]. As a result of this, palatal rugae have become known as the dental equivalent to the fingerprint, unique to every individual (Figure 1).
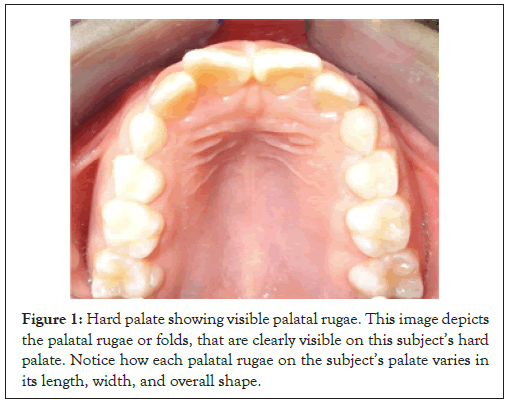
Figure 1: Hard palate showing visible palatal rugae. This image depicts the palatal rugae or folds, that are clearly visible on this subject’s hard palate. Notice how each palatal rugae on the subject’s palate varies in its length, width, and overall shape.
Furthermore, the characteristics of palatal rugae, such as shape, length, or pattern, have been categorized, with significant anatomical differences by gender. In terms of rugae size, 3 categories are present: primary rugae (5-10 mm), secondary rugae (3-5 mm), and fragmentary rugae (<3 mm). Based on shape, palatal rugae have been classified as either straight, wavy (following a sinuous or serpentine figure), curved (crescent-shaped), or circular, with rugae of each category found across different racial ancestries [6]. Thirdly, rugae in the hard palate can either converge, emerging in distinct regions and approaching the same terminal point, or diverge, emerging in distinct regions but extending outwards towards different terminal points (Figure 2). The prevalence of larger palatal rugae, those of primary and secondary classification, is higher in males than in females, with females more likely to have straight patterned rugae [7]. Therefore, palatoscopy (the analysis of palatal rugae) can be established as a potential tool for gender identification, a vital step towards making a positive I.D.
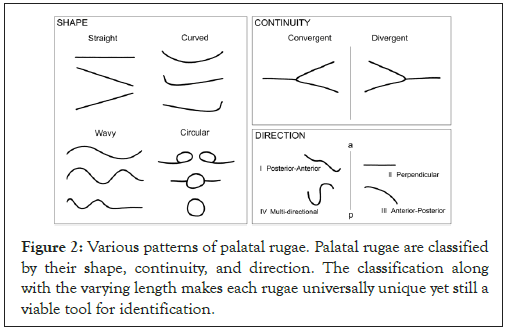
Figure 2: Various patterns of palatal rugae. Palatal rugae are classified by their shape, continuity, and direction. The classification along with the varying length makes each rugae universally unique yet still a viable tool for identification.
Currently, methods of manually analyzing palatal rugae are in place, although not widely used. A 1988 case report by Thomas and Van Wyk utilized dentures found close to a burned remain in order to obtain imprints of the victim’s palatal rugae for comparison – by making plaster casts of the dentures, the lateral and front portions of the anterior palate could be matched. More recently, forensics odontologists at the medicolegal institute of ji-paraná in Brazil received a skeleton remain that had complete dentures; a plaster model of the denture was created, with digital software used to mark the contour of the palate, enabling experts to use marked anatomical similarities in palatal rugae in identifying the victim [8]. Despite these successes, manual determination based on palatal rugae is still time-consuming and expensive, as experts must transform rugae imprints onto a series of molds before visual analysis can begin. Additionally, bias and human error is a possibility in any manual determination, given that localized features of single rugae (such as bends, points of convergence, and inner folds) must be identified before gender identification can occur.
Machine learning provides a tool with which both palatal rugae can be analyzed and human error can be minimized. With machine learning, a computational algorithm builds a model on training data, learning from this data to improve its performance on a given task. In the case of gender identification, a subset of machine learning algorithms known as supervised classification models can be harnessed: these models leverage training data comprised of various features in order to classify input data into a given category, or label. Examples of classification models include K-nearest neighbors, a model in which a data point’s predicted classification is based on the classification of the majority of its “neighbor” data points. Supervised classification models must be trained on cleaned, quantitative data in order to generate proper predictions. The correct label for any given data point must be present to verify the predictions of the model, essentially supervising the behavior of the algorithm to maximize predictive accuracy.
Machine learning is an emerging tool in forensic odontology and has yet to demonstrate its full potential in palatoscopy. While classification algorithms such as the Logistic Regression model (where a prediction is generated based on the probability of a binomial or multinomial event occurring) have been identified as useful techniques in classifying the shape and characteristics of palatal rugae [9], an extensive system for palatoscopy-based gender identification has not been implemented. Indeed, a machine learning model that harnesses the geometric characteristics of an individual’s palatal rugae in performing gender identification can greatly assist forensic examiners. Because palatal rugae patterns are unchanging throughout one’s lifetime, with rugae size stabilizing after childhood, the ability for machine learning to extract patterns in input data, in this case palatal rugae features, can be heavily exploited.
Thus, this project will produce a dynamic pipeline for data collection, processing, and ultimately, gender identification. A dental database of anonymized palate images will be utilized as input data, which must be digitally processed to obtain the geometric features of each individual’s palatal rugae, namely width, length, and overall shape. Using this input data, four classification models will be built: decision tree classifier, random forest classifier, logistic binary classifier, and K-Nearest Neighbors. Our models will be regularly evaluated and will undergo extensive hyperparameter tuning, a system in which an optimal set of model parameters, those that maximize model accuracy and robustness, will be found. The most successful classification model will be selected as part of the Rugae Classification Sequence (RCS), a novel system for gender identification entirely based on the anatomical features of the palatal rugae of a given victim. This system will not only generate classifications far more rapidly than manual determination would, but can run in an automated manner, eliminating much of the human error and bias prevalent in visual forensic examination [10].
The engineering goals for the RCS are as follows: a minimum 60% accuracy and specificity score in gender identification, along with a relatively small runtime per classification generated (indicating low computational intensity). These measures were identified as satisfactory due to the reduced availability of requisite palate images (limiting the possible size of the training set). Furthermore, given that this is a novel application of artificial intelligence to palatoscopy, accuracies around the 60% range provide a substantial baseline for future research that may harness a more comprehensive and targeted palatal rugae dataset [11-15].
The first step in building a machine learning model is to procure requisite data. The primary component of the necessary dataset would be the dimensions of a given palatal rugae, specifically its length and width, taken along the longest and widest parts of the rugae. Obtaining digital measurements from images of palatal rugae is an expeditious task, one that does not use up excess amounts of time or computational resources. The other critical feature would be the age of the patient, in years, since rugae size gradually increases until adulthood. We chose only rugae length, rugae width, and patient age as feature variables to ensure our model was usable in real-time investigations, providing the greatest amount of information regarding rugae characteristics. Additional features such as rugae depth or fold perimeter were not included in the model–it would be impossible to calculate these values for a large set of palatal rugae within a reasonable timespan. The output of the model would be the biological sex of the victim (male, female, or other), rendering this project a classification task [16-18].
We contacted Dr. Malinda Lam, a dentist in the local Puget Sound region, who graciously provided us with a database comprised of 112 images of the mouth. Each image was taken through an intraoral camera and logged through Patterson dental software. The images were completely anonymized, containing only the interior of the patient’s mouth to protect patient confidentiality [19]. We began by performing a preliminary analysis of the database, verifying that at least one rugae measurement could be obtained with a high degree of accuracy from each image. Most photos were focused on the upper palate and adjacent portions of the oral cavity, allowing us to keep every image we obtained. Crucially, as each image was taken from a single patient, the age and gender of the patient was logged in the database [20-22].
Although most of the dental images captured more than one palatal rugae (with the average value being 3.8 rugae per image), many were not clearly visible or adequately defined. Thus, we set down three criteria for all palatal rugae that would be included in the final dataset: direct visibility, definition, and standard characteristics, meaning a lack of external damage or blockage. Once the measurable rugae were identified, we began calculating their dimensions through the digital ruler tool in adobe photoshop. Before taking any digital measurements, the scale of the images was to be set; this would allow Photoshop to automatically convert measurements in pixels to the physical dimensions of the rugae in centimeters. We did this by harnessing the image metadata, which included the length of the patient’s central incisor (from apex to the enamel) for scaling purposes. For each image, before placing the digital ruler, a line was drawn across the length of the central incisor and set to the value (in cm) provided in the metadata, allowing us to obtain physically accurate dimensions from the digital ruler tool [23, 24].
Most rugae are curved or angled, so single, straight rulers would not be effective in capturing the length measurement. Instead, each rugae’s full length was broken down into smaller, straight segments, with photoshop’s ruler tool applied to each segment individually. The sum of all the length segments provided us the total length of a given palatal rugae, or the continuous distance from the initial point of the rugae to the farthest fold of the same rugae, where each fold is defined as a clear rise of skin along the palate. Indeed, many rugae maintained such a high degree of curvature that 7-10 sub-segments had to be individually measured and later summed to pinpoint rugae length [26].
We defined the width of the rugae as the distance between two perimeter points at the widest portion of a given rugae. This ensured that we would be taking the measurement at the widest portion of a given palatal rugae, standardizing the process for each trial performed. Unlike the length measurement, this could be taken through a single, straight digital ruler (Figure 3). For each rugae, the length and width were taken thrice (for 3 trials) using the aforementioned collection procedure–the mean of all 3 trial measurements were the final rugae length and width. In all, from the 112 provided images, 408 rugae were measured, each filling a single row in the training dataset (which was stored as an Excel file). The age and gender of the patient whose rugae were measured were included in the same row, along with the matching image number (from the original database).
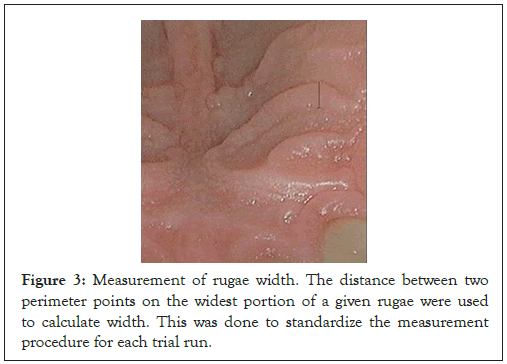
Figure 3: Measurement of rugae width. The distance between two perimeter points on the widest portion of a given rugae were used to calculate width. This was done to standardize the measurement procedure for each trial run.
Python was the coding language of choice due to the availability of machine learning and data analysis libraries, namely Pandas, NumPy, and sklearn. To ready the dataset for statistical modeling, the excel file was converted into a .csv file format and read into Google Collaboratory as a dataframe object. Because classification algorithms require quantitative data, every column had to hold a numerical quantity–we consequently one-hot encoded the patient gender column, mapping the categorical data to corresponding numerical weights. To analyze trends by patient gender and identify anatomical disparities, the dataframe was split into the male rugae dataset and the female rugae dataset. Methods in the NumPy library, such as np.mean(), np.std(), and np.corrcoef() were utilized to calculate measures of central tendency and the correlation between rugae length and width. Further, the matplotlib library was imported to visualize the relationship between gender and rugae size on 2 and 3-D scatter plots. We also attempted to fit the rugae dataset to a number of mathematical transformations, including Power Law (log(x) by log(y)) and Exponential (log(x) by y); this did not yield any improvements to the linearity of the plotted data, however, so the unedited data was maintained [27-29].
The features that would be inputted into the machine learning model would be rugae length (cm), rugae length (width), and age (years), while the output would be the gender classification. Following statistical analysis, the overall dataset was split into the feature variables (matrix X) and the labels (vector y). Next, both the feature matrix and label vector underwent an 80-20 split for training and testing respectively; that is, 80% of the data points (one per palatal rugae) were set aside to train a given machine learning model, while 20% of the data points were devoted to testing the model. This split is fairly common in research and provides the model enough “experience” with which to maximize predictive accuracy. The testing dataset enables us to check the model’s robustness and accuracy on predictions for unknown patients: the model is not exposed to any testing data points beforehand and instead must rely on developed mathematical patterns in order to generate classifications.
Four supervised machine learning classification algorithms were applied in a novel way to the refined dataset for the purposes of gender classification based on anatomical characteristics of the palatal rugae. The algorithms used are as follows: Decision Tree Classifier, Random Forest Classifier, Binary Logistic Classification, and K-nearest Neighbors. A Decision Tree Classifier is a “tree” of nodes connected with branches, each representing a single decision, or condition, that a data point may satisfy. The nodes closest to the bottom are known as leaves, displaying the final possible classifications. Random Forest Classifiers are simply an ensemble of decision trees – the algorithm compiles every prediction generated by the ensemble, with the “majority vote” serving as the final classification. Binary Logistic Classification models rely more on mathematical functions, namely the sigmoid function, which uses parameters in the form of linear equations in generating a probability value between 0 and 1–if the probability is greater than 0.5, the model outputs “True”, whereas a probability below 0.5 results in a “False”. The fourth and final algorithm we harnessed was the K-nearest Neighbors model, which plots every data point on a multi-dimensional graph, calculating the distance between the test data and the training data to select the K points closest to the test data. The model outputs the classification held by the majority of these neighbors, similar to the majority vote performed in Random Forest Classifier. The aforementioned machine learning algorithms are all available through the sklearn library, which was imported to our Google Collaboratory file prior to applying any models to the data.
Indeed, the task of calling each algorithm and fitting it to training data was quite simple due to the practicality of the sklearn library, which contains methods such as .fit() and .predict() to facilitate fitting and testing of various models. It would be more significant to discuss the extensive hyper parameter tuning applied to every model following the initial testing-with regards to artificial intelligence, tuning refers to the adjustment of various hyperparameters that dictate the behavior of a given model, including learning speed, depth, and other modelspecific characteristics. Both sklearn’s GridSearchCV program and sequential iterative tuning were tested and evaluated in order to maximize predictive accuracy. GridSearchCV essentially uses a grid of randomly selected values assigned to a hyperparameter, finding the optimal combination of values from this grid – while this program relies on randomness, it is computationally efficient, as only small subsets of values are tested. However, because of the limited training size, which forces the models to learn faster and glean more from each individual data point, iterative sequencing, where the range of possible parameter values can be manually decided, was selected as the method of choice.
The first constructed model, Decision Tree Classifier, had the following parameters tuned: random_state (seed value for randomness with which input data is shuffled), max_depth (maximum height of the tree), min_weight_fraction_leaf (minimum proportion of the sum of all weights required to be at every leaf node), and min_ impurity_decrease (minimum impurity in predictive power before a node is “split” into two children). For each hyperparameter, the code iterated through 50-100 potential values–each value was plugged into a separate Decision Tree model that was trained and tested on the rugae dataset. The program stored the value that maximized model f1-score, providing it as output following the iteration. Although the Decision Tree class (and most other ML algorithms) contains many more hyperparameters, only the aforementioned parameters were tuned, as they produce the greatest impact on model behavior and are applicable to most datasets. This procedure was repeated for the three other algorithms used, although with varying parameters: the Random Forest algorithm had the same hyperparameters tuned as the Decision Tree Classifier, since the former is simply an ensemble of the latter. We chose to set the parameter Bootstrapping to True for the Random Forest algorithm, as this would allow each Decision Tree within the ensemble to receive a randomly selected segment of the dataset, reducing the chances of general over fitting and thus improving predictive accuracy (Figure 4).
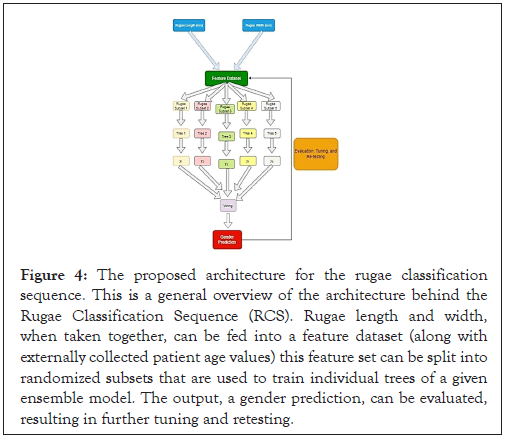
Figure 4: The proposed architecture for the rugae classification sequence. This is a general overview of the architecture behind the Rugae Classification Sequence (RCS). Rugae length and width, when taken together, can be fed into a feature dataset (along with externally collected patient age values) this feature set can be split into randomized subsets that are used to train individual trees of a given ensemble model. The output, a gender prediction, can be evaluated, resulting in further tuning and retesting.
For the Logistic Binary Classifier model, we tuned the numerical parameters max_iter, which handles the maximum number of iterations the model can make over the training set before converging, and tolerance, which dictates the maximum amount of error in the model that can be tolerated before stopping. The penalty and solver parameters were set to “l2” and “saga” respectively l2 penalizes the model for having excessive sensitivity to a single data point, while the saga solver significantly improves runtime for both large and small datasets. Finally, we tuned the n_neighbors and n_jobs parameters for the K-nearest Neighbor algorithm-n_neighbors dictates the number of adjacent data points taken into consideration when generating a classification (usually an odd number >1), while n_jobs enables the model to run a specific number of parallel operations during neighbor detection. We additionally set the algorithm type to “brute”, which brute-force searches for every neighbor, optimal for smaller datasets such as ours (with only 408 data points). This concluded hyperparameter tuning and allowed us to begin running the finalized models.
The Random Forest, Decision Tree, Logistic Binary Classifier, and K-nearest Neighbor models with the updated hyperparameters were trained on 80% of the rugae dataset and tested on the separate, unseen 20%. For each model, a confusion matrix was generated to visualize the precision, accuracy, recall, and general f1-score, the equations of which are provided below. Note that the one-hot encoding procedure assigned a 0 for male victims (False, or negative) and a 1 to female victims (True, or positive).
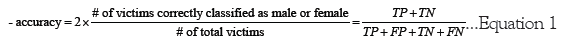
Where in the accuracy is equal to the frequency of correct classifications divided by the total number of victims in the dataset (note that TP is true positive, FP is false positive, TN is true negative, and FN is false negative).
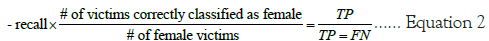
Where in the precision is equal to the frequency of correctly identified positives (females) divided by the total number of positive predictions.

Where in the recall is equal to the frequency of correctly identified positives (females) divided by the total number of female victims.
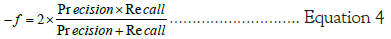
Where in a more representative metric of overall model accuracy is determined as a score from 0 to 1 (the higher the more effective). Each model’s accuracy measures were compared, with the highest-achieving model labeled a component of our novel Rugae Classification Sequence (RCS). The results of each model were broken down by gender to analyze disparities in predictive accuracy, further establishing the role gender has in dictating the anatomical characteristics of the palatal rugae [30].
Statistical analysis was first conducted for all patients in the dataset; the mean palatal rugae length for all subjects was 0.748, with females having a mean rugae length of 0.716 cm and males having a greater mean rugae length of 0.778 cm (Figure 5). The mean palatal rugae width for all subjects was 0.161 cm, with negligible difference between male and female patient groups. We also found the correlation coefficient r between palatal rugae length and width to be 0.305; further, the correlation coefficient between rugae length and width was significantly higher for female palatal rugae than for male palatal rugae (0.391 versus a minimal 0.163) (Table 1). Regarding the relationship between age and characteristics of the palatal rugae, the correlation coefficient between age and rugae length was found to be a minimal 0.028, while the correlation coefficient between age and rugae width was -0.106 (this may be due to the ages of the patients sampled, a limitation that will be elaborated on in Section 4) (Figure 6).
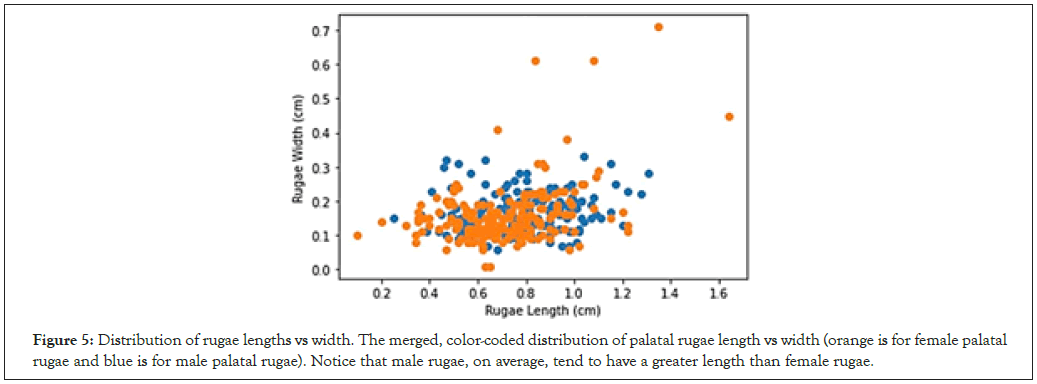
Figure 5: Distribution of rugae lengths vs width. The merged, color-coded distribution of palatal rugae length vs width (orange is for female palatal rugae and blue is for male palatal rugae). Notice that male rugae, on average, tend to have a greater length than female rugae.
| Gender | Mean length (cm) | Standard deviation length (cm) | Mean width (cm) |
Standard deviation width (cm) |
Correlation coefficient (between palatal rugae length and width) |
|---|---|---|---|---|---|
| Female | 0.716 | 0.214 | 0.160 | 0.160 | 0.391 |
| Male | 0.778 | 0.183 | 0.163 | 0.053 | 0.163 |
| All | 0.748 | 0.201 | 0.161 | 0.071 | 0.305 |
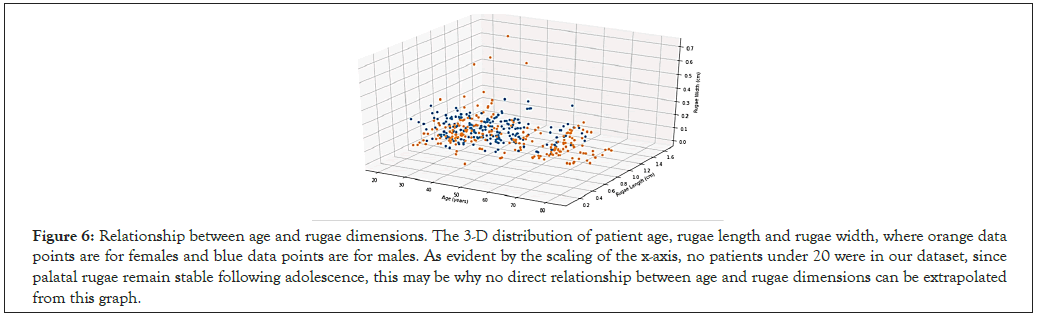
Figure 6: Relationship between age and rugae dimensions. The 3-D distribution of patient age, rugae length and rugae width, where orange data points are for females and blue data points are for males. As evident by the scaling of the x-axis, no patients under 20 were in our dataset, since palatal rugae remain stable following adolescence, this may be why no direct relationship between age and rugae dimensions can be extrapolated from this graph.
The accuracy of gender classification varied by model tested. The Decision Tree Classifier achieved the lowest f1-score of 0.522, with a general accuracy of 59.8% on 82 samples tested (Figure 7). In contrast, the K-nearest Neighbor model (with optimal hyperparameters n_ neighbors=41 and algorithm=‘auto’) achieved the highest f1-score and accuracy at 0.592 and 64.6% respectively on the same 82 samples. A specificity score of 0.667 confirmed that K-nearest Neighbors was a relatively effective predictive algorithm (Figure 8). Moreover, our Random Forest model achieved lower accuracy scores, with an f1-score of 0.531 and an accuracy of 63.4% (Figure 9). Interestingly, while the Logistic Binary Classifier obtained an f1-score of 0.569, its minimal precision and accuracy scores (at 0.442 and 0.524 respectively) hinted at over prediction (Figure 10). Due to the small number of features and data points, the final runtime for each model remained below 1.5 seconds, satisfying requirements set for computational efficiency. In all, the K-nearest Neighbors model was set as the primary machine learning model for the novel RCS (Table 2).
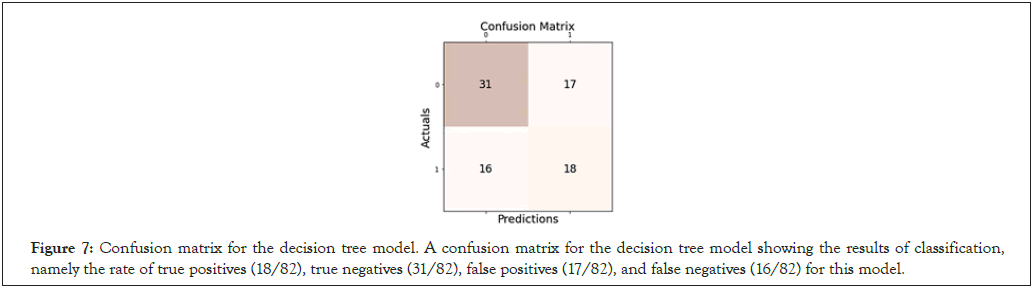
Figure 7: Confusion matrix for the decision tree model. A confusion matrix for the decision tree model showing the results of classification, namely the rate of true positives (18/82), true negatives (31/82), false positives (17/82), and false negatives (16/82) for this model.
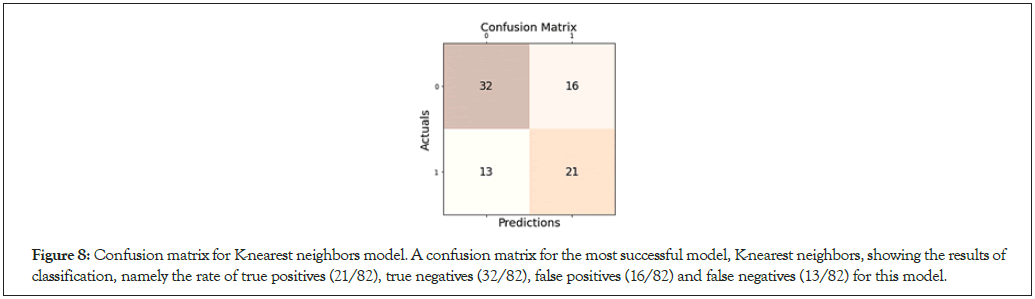
Figure 8: Confusion matrix for K-nearest neighbors model. A confusion matrix for the most successful model, K-nearest neighbors, showing the results of classification, namely the rate of true positives (21/82), true negatives (32/82), false positives (16/82) and false negatives (13/82) for this model.
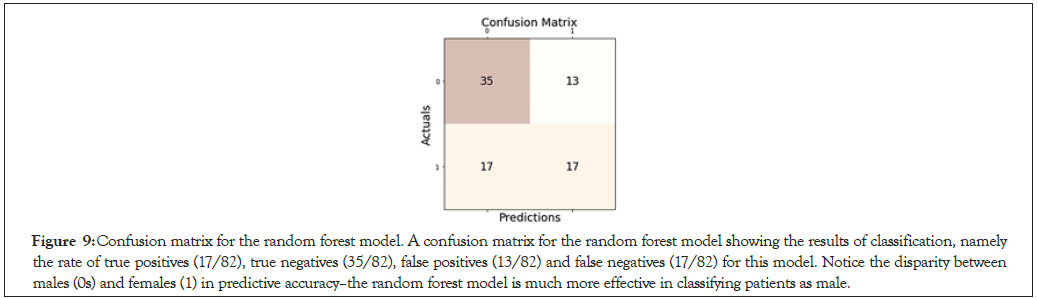
Figure 9: Confusion matrix for the random forest model. A confusion matrix for the random forest model showing the results of classification, namely the rate of true positives (17/82), true negatives (35/82), false positives (13/82) and false negatives (17/82) for this model. Notice the disparity between males (0s) and females (1) in predictive accuracy–the random forest model is much more effective in classifying patients as male.
| Algorithm | Precision | Accuracy | F1-Score | Recall |
|---|---|---|---|---|
| Decision tree | 0.514 | 0.598 | 0.522 | 0.529 |
| Random forest | 0.567 | 0.634 | 0.531 | 0.500 |
| Logistic binary | 0.442 | 0.524 | 0.494 | 0.559 |
| K-nearest | 0.568 | 0.646 | 0.592 | 0.618 |
Our analysis of the dataset clearly demonstrates a disparity between male and female palatal rugae characteristics. Rugae found in male patients tend to be longer (0.748 cm for males vs. 0.716 cm for females), although rugae width is too variable to exhibit significant difference between genders. Moreover, our study shows that palatal rugae maintain no shared uniform structure; given the low calculated correlation coefficient of 0.305 between palatal rugae length and width, the shape of palatal rugae varies greatly. This was confirmed by our visual analysis of the dental database, where rugae of many forms were found, ranging from curved to wavy. Notably, female palatal rugae exhibited a significantly higher correlation coefficient between rugae length and width than male rugae (0.391 for females vs. 0.163 for males)–female rugae may simply follow a more predictable structure due to their shorter length, where rugae of larger length tend to have larger widths.
In terms of the relationship between age and anatomical characteristics of the palatal rugae, the remarkably low correlation coefficient values between age and rugae length/width (0.028 and -0.106 respectively) may be due to limitations in the dental database. Because the dental firm providing the images mainly operated on adult patients, all entries in the database were from individuals aged 25 to 80. As established previously, palatal rugae remain stable following adolescence, with little to no growth or development from this period until death. Because of this stability, no trends can be extracted from the dataset between a patient’s age and the size of their palatal rugae. Future studies could include children and adolescents in order to study the growth of palatal rugae over this period and further establish the importance of age in dental development. Moreover, the relationship between gender and characteristics palatal rugae may differ significantly between adults and children, so a more generalized analysis would address this shortcoming of our study.
The machine learning models constructed to predict the gender of a victim given dimensions of their palatal rugae, when evaluated together, satisfied engineering goals for accuracy and computational efficiency. The Decision Tree Classifier and Logistic Binary Classifier performed quite poorly on the dataset, with the latter heavily overpredicting the occurrence of female victims. A high recall score of 0.853 and an opposingly low precisions score of 0.426 further demonstrate that the Logistic Binary Classifier did overpredict, simply marking most data points as female, or positive. The most successful model, K-nearest Neighbors, achieved the highest accuracy score (0.65) and specificity score (0.68). Because this is greater than the required accuracy score of 0.6, we are able to set the K-nearest Neighbors (KNN) algorithm as the primary component of our RCS. Regarding computational requirements, KNN took under 1.5 seconds per prediction, far faster than most forensic experts would take in the field to collect, measure, process, and analyze a victim’s dental records. Mass disasters require efficient forensic analysis, and the RCS provides the needed efficiency and relative precision in determining general characteristics of a given victim.
Machine learning is a novel technique in palatoscopy; our KNN model, as part of the RCS, demonstrates that machine learning can be successfully applied to dental records. No other studies, to our knowledge, harness AI for the purpose of forensic palatoscopy, rendering the RCS a novel, emerging tool for rapid identification. Although accuracy scores are mid-tier (just under 70% for most tests), they have the potential to increase dramatically given a more substantial database. Palatal rugae datasets are sparse, with our dataset derived from a dental database following significant processing; this meant that scaling issues and visual obstructions could introduce sizeable error to our measurements. Future studies can take direct, focused images of the palate, providing more accurate measurements of rugae length and width–as a result, a clearer relationship between gender and rugae size can be extracted through machine learning, improving accuracy and precision of any given model. Furthermore, the procedure used to perform hyperparameter tuning, which increased accuracies by up to 10% for the KNN algorithm, could be refined in future works; instead of iterative sequencing, which introduces the chance of overprediction, the randomized grid technique (which was not attempted due to constraints in dataset size) could greatly reduce model training time and inculcate the models with more balanced parameter values (e.g., those that increase both precision and accuracy/recall).
The RCS is a novel application of machine learning in palatoscopy, and with these results, has the potential for large-scale application–indeed, it may provide forensics experts with a more efficient and reliable tool to identify victims given dental remains. Our model can be integrated with future age regression models–those that predict a victim’s age from dental features–to provide an end-to-end system for victim identification. Other datasets focused on quantifiable anatomical characteristics can utilize and adjust our classification model to predict gender, as sexual dimorphism applies to a wide swathe of human anatomy. In all, the RCS is a novel application of machine learning in forensic palatoscopy, modeling the relationship between gender and palatal anatomy.
We would like to thank Dr. Malinda Lam, who provided us with her expertise in dental anatomy.
None to report.
The authors received no financial support for the research, authorship and/or publication of this article.
Institutional review and clinical trial information not applicable.
Citation: Singh A, Gershony S (2022) The Rugae Classification Sequence: Using Machine Learning to Determine Gender from Characteristics of Palatal Rugae. J Odontol. 6:629.
Received: 08-Aug-2022, Manuscript No. JOY-22-18725; Editor assigned: 11-Aug-2022, Pre QC No. JOY-22-18725 (PQ); Reviewed: 25-Aug-2022, QC No. JOY-22-18725; Revised: 01-Sep-2022, Manuscript No. JOY-22-18725 (R); Published: 09-Sep-2022 , DOI: 10.35248/JOY.22.6.629
Copyright: © 2022 Singh A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.