Research Article - (2019)Volume 5, Issue 2
Towards Managing and Controlling Aflatoxin Producers within Aspergillus Species in Infested Rice Grains Collected from Local Market in Kenya
Youmma Douksouna1*, Andrew Nyerere1,2, Joel Masanga3, Steven Runo3 and Zachée Ambang4Abstract
Rice grain can be attacked by a range of pathogens, including Aspergillus species, which can cause accumulation of aflatoxins that represent a serious threat the consumers. Aflatoxins are naturally occurring toxic metabolites synthesized by certain species of Aspergillus. This study was designed to analyze the prevalence of Aspergillus species and aflatoxin-producing Aspergillus using polymerase chain reaction (PCR) in rice grains being sold in the local markets. A total of 98 samples were randomly collected and primarily analyzed to observe the moisture content and fungal growth. Subsequently, Aspergillus species were isolated, characterized using ITS primers and screened for aflatoxigenic fungal targeting specific genes (nor-1 and ver-1) involved in aflatoxin biosynthetic pathway using PCR assay. It was observed that all tested samples were found contaminated with the highest prevalence of Aspergillus species and aflatoxigenic fungal, 55.4% and 36.4% for nor-1 and ver-1 respectively. Occurrence of high contamination level of Aspergillus species indicates the possible production of aflatoxins in rice grains. A total of 74 genomic DNA extracted from the isolates in this study, 55.4% of isolates were confirmed the aflatoxin producers by targeted genes. This research provides the baseline studies for the occurrence of Mycotoxigenic fungal species in rice grains being sold in local markets.
Keywords
Rice grains; Aspergillus species; Prevalence; ITS; Nor-1; Ver-1; Aflatoxigenic
Introduction
Rice is a cereal consumed by great part of the human population throughout the world in many forms of products, such as white rice, parboiled rice, meal rice and rice bran [1]. After harvesting, it is generally dried and under inappropriate storage conditions such as storage dumpy/misty, lack of controlling airborne, insects, temperature and humidity during storage, rice is considered a viable substrate for growth of fungi [2,3]. During processing, rice kernels can be contaminated with fungi especially those producing mycotoxins such as aflatoxins given the conditions are favorable [4]. Additionally, delayed drying process and excess moisture (above 13%) [5] can promote the growth of fungi. Several strains of Aspergillus section Flavi are common in plants and its processed derivatives, with some producing diverse mycotoxins, such as aflatoxins, 3-nitropropionic acid, tenuazonic acid and cyclopiazonic acid [6]. The production of aflatoxins is associated with spore production by species of Aspergillus [7]. They are produced as secondary metabolites and 4 types i.e., aflatoxin B1 (AFB1), B2 (AFB2), G1 (AFG1) and G2 (AFG2) are the most important with reference to carcinogenic effect, mutagenic, teratogenic, and immunosuppressive capabilities [8]. They are classified in group I as first class carcinogens, mutagens and immunosuppressive agents by International Agency for Research on Cancer [9]. About 5.2 million cancer deaths occur each year, 55% of which occur in developing countries [10]. The removal of AFS is difficult due to their stability and thermal resistance in dried products [11]. In fact, AFS are resistant to food processing and thus they may remain throughout the food chain [12]. Therefore, AFS are potential threats to human health, either by consumption of direct contaminated food products or by carry over aflatoxins and their metabolites in milk and meat [13]. World health authorities warn that low doses with long-term dietary exposure toaflatoxins are a major risk to lead hepatocellular carcinoma [14]. In this context, the aim of this study was to characterize the nature of Aspergillus species contaminating rice grains on one hand and the aflatoxigenic genes profile of Aspergillus species isolated by using the phylogenetic analysis of Internally Transcribed Spacer (ITS) regions on the other hand.
Materials and Methods
Samples
Samples comprising local and imported rice grains were collected randomly (1 kg) from local retail markets and millers in Mwea and Thika (Kenya) and labelled appropriately. A total of 98 samples (local rice produced in Mwea and imported rice originating from Biriyani, India, Pakistan, Thailand) were taken according to the alternative sampling plan for official control of mycotoxins in food [15]. To give a representative sample which was then put in sealed bags and transported to the Molecular Biology and Biotechnology Laboratory in PAUSTI.
Moisture content analysis
To better understand the factors that may have led to aflatoxins contamination of the samples, each sample of rice grains collected was analyzed for its moisture content using Grainer meter (Model Japonica).
Isolation and enumeration of fungal species
Isolation of microflora from the grains was done following Ulster method [16]. The samples were analyzed by the direct plating technique. For each sample, 20 particles (rice grains suspected are contaminated by fungi) were placed aseptically on a layer of moistened filter paper in the Petri dishes. The Petri dishes were incubated in an upright position at 25°C for 5-7 days in darkness and ventilated for 12 hours in the 3rd day. After incubation, the plates were examined and the number of contaminated particles were counted and reported as percentages. Fungi growing on different seeds were isolated from emerging colonies on modified rose Bengal chloramphenicol agar (mRBA) [17]. Pure cultures were carried out for subsequent studies using two (2) culture media: malt extract agar (MEA) and potato dextrose agar (PDA) according to Pitt et al. [18] and Varga et al. [19] for seven days.
Phenotypic characterization
Phenotypic characterization was done on the basis of mycelium growth pattern, color and properties of fruiting bodies of the fungi. Growth pattern of fungal colonies, colony size (diameter) and color (reverse and coarse), exudates and colony margins were the macroscopic features used in identification. Size of head, vesicle shape, phailides, matulae, conidiophores and conidia (conidial diameter, wall, shape, and surface and conidia attachment with condiophore) were done for microscopic identification. These features were finally compared with the synoptic keys for identification of the isolated fungi [20].
Percentage occurrence of each species isolated was also calculated
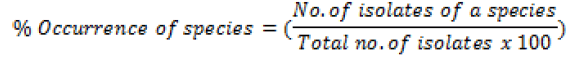
Molecular characterization of the isolates
The genomic DNA was extracted from seven days’ old fungi cultures grown on MEA and PDA media and the fungal mass from the pure cultures was scraped-out from the plates. 50-100 mg of fungal mycelium was scraped and placed in a 2 ml tube and vortexed vigorously for 30 minutes with glass beads to crush the mycelial wall and to release the DNA. A volume of 500 μl of Lysis Buffer (100 mM Tris-HCl pH 8, 1 mM EDTA, 100 mM NaCl, 10 mM B-mercaptoethanol and 1% Sodium dodecyl sulfate), 5 μl of RNase A and 1 μl of Proteinase K were added. Tubes were then incubated at 65˚C for 45 minutes into buffer.
Thereafter, 270 μl of Sodium/Potassium acetate (3 M) was added and samples were well mixed and centrifuged at 13000 rpm for 10 minutes. After centrifugation supernatant (700 μl) was transferred to a fresh tube, and an equal volume of Chloroform: Isoamylalcohol (24:1) was added and mixed well, samples were stood on bench for 5 mn followed by centrifugation at 13,000 rpm for 10 min. The supernatant (700 μl) was transferred into new tubes, 80 μl of Sodium/Potassium acetate (3 M) and 587 of ice cold Isopropanol were added then mixed well by inverting the tubes then incubate at -20˚C overnight. After that, samples were centrifuged at 13,000 rpm for 30 min, then supernatant was discarded carefully. The DNA pellets were washed with 1 ml of 70% ethanol and centrifuged at 13,000 rpm for 10 min. The DNA pellets were air dried and dissolved in 50 μL of TE buffer and stored at -20°C until use.
Separation of the isolated genomic DNA was done using agarose gel electrophoresis followed by Sybr Green visualization using Lambda (λ) as DNA size marker. The concentration and purity of genomic DNA were measured using Nano-drop Spectrophotometer (Model PCR Max Lambda), purity of all extracted DNA was done by taking their absorbance at 260 nm and 280 nm.
Diagnostic PCR using Aspergillus universal primers
We used Aspergillus universal primer pairs to amplify the ITS1 and ITS4 region of different isolated Aspergillus strains for characterization and primer sequences already available in literature [21] to amplify 2 different genes involved in Aflatoxin biosynthetic pathway Norsolorinic Acid (NOR): aflD (nor-1), and Versicolorin: aflM (ver-1) fragments of aflatoxigenic fungal genomic DNA. The sequences of primers are listed in Table 1.
| Set | Primer name | Sequences (5’-3’) | Length of PCR Product (bp) |
|---|---|---|---|
| 1 | ITS1 | F-TCCGTAGGTGAACCTGCGG | 598 |
| ITS4 | R-TCCTCCGCTTATTGATATGC | ||
| 2 | aflD | F-ACCGCTACGCCGGCACTCTCGGCAC | 400 |
| R-GTTGGCCGCCAGCTTCGACACTCCG | |||
| 3 | aflM | F-GCCGCAGGCCGCGGAGAAAGTGGT | 536 |
| R-GGGGATATACTCCCGCGACACAGCC |
Abbreviations: F-Forward; R-Reverse
Table 1: Oligonucleotide primer sets used for the study.
PCR assay was performed in 20 μL of a reaction mixture that contained 1 μL of extracted genomic DNA, 4 μL of master mix, 0.5 μL of each forward and reverse primer. The final volume was made up to 20 μL with nuclease free water. Amplification was performed in a Proflex PCR System (Model 4483636) with the following conditions (Table 2).
| S.No | PCR fragment | Initial denaturation | Denaturation | Annealing | Elongation | Final elongation |
Number of cycles |
|---|---|---|---|---|---|---|---|
| 1 | ITS | 94°C 5 mn | 95°C 60 s | 52°C 60 s | 72°C 60 s | 72°C 10 min | 35 |
| 2 | aflD | 94°C 5 mn | 94°C 60 s | 64°C 60 s | 72°C 60 s | 72°C 10 min | 33 |
| 3 | aflM | 95°C 5 mn | 95°C 60 s | 65°C 60 s | 72°C 2 mn | 72°C 10 min | 33 |
Table 2: PCR parameters of the oligonucleotide primer sets used for the study.
The amplified PCR products were resolved by gel electrophoresis in a 1.5% agarose (Sigma) gel stained in 10 μL of Trugel Fluorescent Dye. The DNA bands resolved on agarose gel were visualized in UV Documentation system with camera (Model UV Doc. HDS UITEC Cambridge) transilluminator. The sizes of the amplicon were estimated by comparing with a commercial 1 kb DNA ladder on agarose gel.
Results
Moisture content
In present study mycoflora was isolated from rice grains to evaluate the contamination level in rice grains sold in the market. Samples collected were previously examined for moisture content and the prevalence of Aspergillus species. Fungal growth in food samples is usually influenced by various factors. Climatic conditions, especially temperature and higher moisture content of the food samples play an important role in this process. All samples were initially analyzed for moisture content to check for the relationship correlation (Y=4.211* × -54.39) between moisture level in rice grains and fungal growth. 4 groups (group 1, group 2, group 3 and group 4) of samples were clustered according to their percentage 14-14.5; 15-15.5; 15.6-16.5; and 16.6-17.9 respectively as shown in Figure 1.
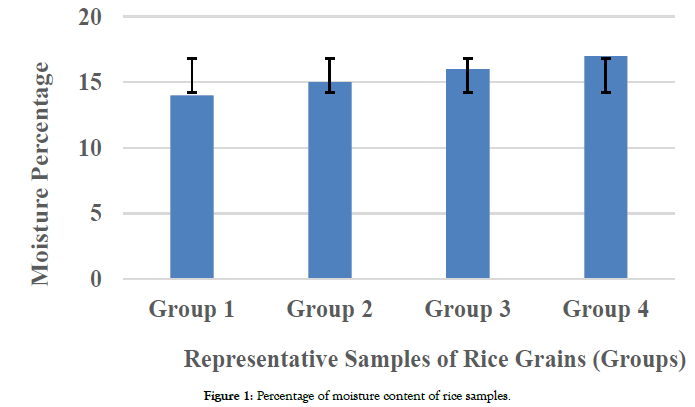
Figure 1. Percentage of moisture content of rice samples.
Samples with high moisture contents for a length of time offer best microclimate for the growth of mycotoxigenic fungi and mycotoxins production. A positive correlation between moisture content and isolates was observed in this study: R2=0.2926, with the increase of moisture content the number of isolates was increased, indicating a higher level of fungal growth and contamination of rice grains.
Isolation of Aspergillus species
The results of isolates for different samples showed the presence of Aspergillus in all samples. From the observations made in infested rice grains the fungal growth was seen in all the inoculated plates except negative control.
Morphological analysis
All the samples were positive for Aspergillus with a total of eight (8) fungal strains including. Aspergillus clavatus, Aspergillus flavus, Aspergillus fumigatus, Aspergillus nomius, Aspergillus oryzae, Aspergillus parasiticus, Aspergillus versicolor, and Aspergillus spp being observed. These strains were purified based on their distinct morphology on malt extract agar (MEA) and potato dextrose agar (PDA) plates and differentiated based on their macroscopic and microscopic characteristics. Macroscopic identification was based on colony and reverse color, diameter, exudates and texture. On the basis of macroscopic study, isolates can’t be differentiated. Further microscopic study involved the arrangement, color, diameter, shape, size, wall characters, cellular contents, conidial heads, conidiophore, sterigmeta, conidia and conidial arrangements was observed in microscope [22]. As the macroscopy and microscopy characteristics of fungal isolates that are presented in Figure 2.
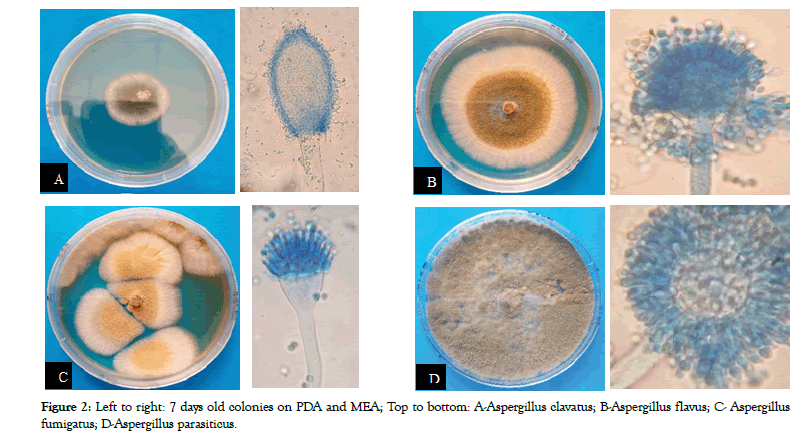
Figure 2. Left to right: 7 days old colonies on PDA and MEA; Top to bottom: A-Aspergillus clavatus; B-Aspergillus flavus; C- Aspergillus
fumigatus; D-Aspergillus parasiticus.
Morphological analysis revealed that Aspergillus flavus (37.5%) was the most represented strain followed by Aspergillus parasiticus (27.9%), Aspergillus fumigatus (10.8%), Aspergillus Nomius (6.2%), Aspergillus oryzae (5%), Aspergillus Clavatus (3.1%), Aspergillus Versicolor (2.7%), and Aspergillus spp (6.5%). The ability of fungi to change their morphology at different stages of growth and reproduction renders phenotypic characterization inefficient. Therefore, these isolates were subjected to molecular analysis through PCR amplification of their Internally Transcribed Spacer (ITS) regions and 2 genes profile of Aspergillus species involved in aflatoxin biosynthetic pathway Norsolorinic Acid (NOR): aflD (nor-1), and Versicolorin: aflM (ver-1) fragments of aflatoxigenic fungal genomic DNA Figure 3.
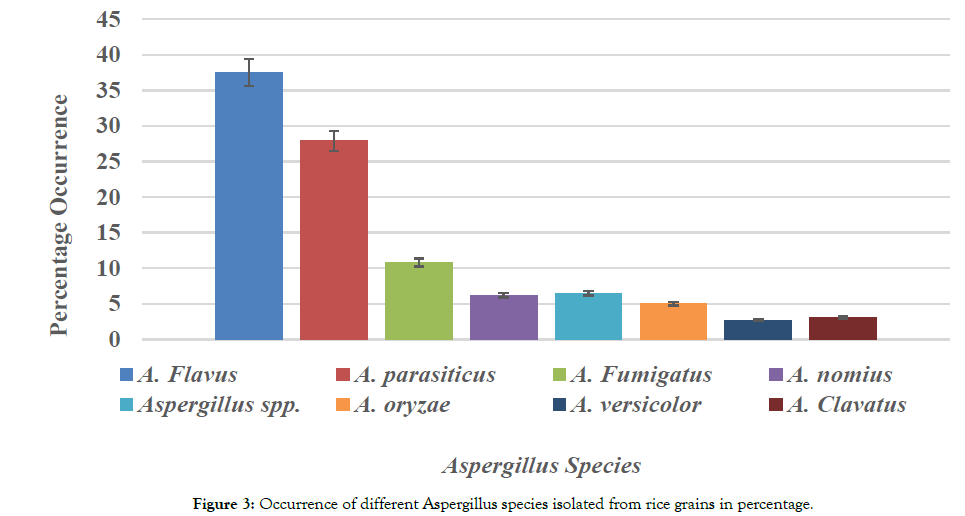
Figure 3. Occurrence of different Aspergillus species isolated from rice grains in percentage.
Molecular characterization
Seventy-four (74) genomic DNA isolated were selected from Aspergillus strains for molecular analysis. Quantity of extracted DNA from all the samples was between 369 μg/mL to 1998 μg/ mL, which is an excellent quantity range. Genomic DNA isolated was amplified through PCR according to the described conditions. The PCR was conducted using the sets of three primers specifically ITS1-ITS4 for characterization of Aspergillus species, aflD and aflM genes profile targeted DNA for screening aflatoxigenic strains from Aspergillus isolates. The expected size of each primer pair produced a single DNA fragment is 598 bp for ITS, 400 and 536 bp for aflD and aflM respectively. From a total of 74 phenotypically characterized Aspergillus species isolates, all samples showed positive PCR results for ITS primer pair. Although ITS1-ITS4 is the most commonly used primer pair for the detection of Aspergillus species [23], it does not have sufficient discriminative power to differentiate between aflatoxigenic and non-aflatoxigenic (atoxigenic) species. Therefore, primers pair for specific genes profile involved in aflatoxin biosynthetic pathway aflD and aflM were amplified. Altogether, 74 samples were screened in which, 49 samples were found positive to targeted gene aflD, and 27 samples to targeted gene aflM. The amplified products were separated by Agarose gel electrophoresis as shown in Figure 4.
Figure 4. PCR amplicons of A: 400bp product of aflD; B: 536bp product of aflM markers; M: Molecular Weight Marker 1kb (Solis Biodyne); -ve:
Negative Control. Numbers are the code of samples.
Discussion
Moisture content is one of the determining factors for the growth of Aspergillus fungi. Here, high moisture content (14 to 17.9%) was recorded. This is way above of the standard levels of 12-13% for rice grains as reported by Magan et al. [24]. These results are in line with those reported by Hina et al. [25] who attributed the same to the processing, wet weather and transport conditions.
The prevalence of Aspergillus species was analyzed in all samples under study (Figure 1). The investigation of occurrence fungal species showed that all the samples were contaminated. Aspergillus flavus was the most predominant followed by Aspergillus parasiticus as compared to other Aspergillus strains. These dominances could be attributed to the ability of Aspergillus to growth on substrates in a wide range of environment and the production of spores that remain viable even under extremely strict conditions. Our present data (Figure 3) are in agreement with that recorded by several investigators [25-27] who revealed the high prevalence of Aspergillus species in contamination of rice grains from local markets. In that case, it might be due to the rice grain processing, transport and storage conditions such as improper packaging, poor ventilation and increased humidity in the ware houses that favor the growth of storage fungi i.e., Aspergillus species, instead of field fungi i.e., Fusarium species, more frequently found in field rather than on processed and stored grains [28]. The highest prevalence of Aspergillus species detected from different samples, which are the main contaminant of rice grains in present study. The presence of fungal growth indicates the anticipated contamination of mycotoxins; as high prevalence of Aspergillus species indicates the contamination of aflatoxins in rice grains as reported by Aiden et al. [29]. Due to the toxic and carcinogenic properties of aflatoxins, there is an urgent need to develop sensitive, rapid, and specific technique for the identification of aflatoxin producing from food samples. In our study the PCR reaction was targeted against ITS1- ITS4 for detection of Aspergillus species, aflD and aflM genes profile targeted DNA for screening aflatoxigenic strains from Aspergillus isolates. The primers ITS1 and IS4 were previously amplified successfully all the ITS region isolates selected using conventional PCR. In gel electrophoresis of PCR product and amplicon corresponding to 598 bp in size was seen only in positive sample, which clearly indicated that the primers (ITS1-ITS4) were specific for Aspergillus species. This result is in concordance with that recorded of many investigators [23,30] that ITS1-ITS4 is the most commonly used primer pair for the detection of Aspergillus species.
On one hand, these fungi deteriorate the quality of food products and on the other hand, they are able of producing harmful toxins such as aflatoxins. Biosynthetic pathway comprises of many enzymatic steps with aflatoxins as the end product. Several papers have reported use of PCR technology as rapid and sensitive method for detection and diagnosis of aflatoxin production [31] and to detect aflatoxigenic strain from non-aflatoxigenic strain [32,33] in food. During the current study, PCR was performed for the detection of aflatoxigenic genes i.e., aflD (nor-1), and aflM (ver- 1), respectively by using specific designed primers. Cluster genes in aflatoxin biosynthesis pathway contain structural genes, nor-1 and ver-1 are the genes that play a key role in the production of aflatoxins. In our PCR analysis, screening of aflatoxigenic genes profile of isolates revealed that 41 out of 74 isolates were aflatoxin producers for aflD. In this study it was observed that aflD gene was being amplified efficiently as it was a structural gene required in the initial step of aflatoxin biosynthesis pathway. However, aflM primers were less efficient with 27 isolates amplified for this gene. These two primers were carefully selected to be highly specific for these two genes known to be essential for aflatoxin biosynthesis. Each primer pair yielded a single DNA fragment of the expected size of 400 and 536 bp for nor-1 and ver-1, respectively. A total of 74 genomic DNA isolated, only 33 showed negative PCR results for these genes targeted. This result indicates these isolates are non aflatoxigenic. Non aflatoxigenicity of these isolates may be because of a mutation or gene deletion in one or more genes belonging to the biosynthetic gene cluster [34]. Houshyarfard et al. reported that production of aflatoxin is bound to several factors [35]. Firstly, the presence of certain genes, Secondly, the genes should be intact. Meaning that, there should not be major deletions or insertions within the gene regions or regions flanking the gene. Otherwise, deletions of several portions of the aflatoxin biosynthesis gene cluster have been reported to be the main cause for the lack of aflatoxin production. Keeping in view the above results, it is obvious that the contamination level of mycotoxigenic fungi in rice grains sold in the markets is very high. Our results are in accordance with other reports demonstrated that the significance of PCR based techniques to detect aflatoxigenic potential of Aspergillus strains [6,21,26,27,36,37].
Conclusion
On the basis of achieved results, we can say that prevalence of Aspergillus species in rice grains sold in the markets which is very high, and indicates the possible high level of aflatoxigenic strains in rice grains under study. In this study, 55.4% of isolates were able to confirm the aflatoxin production by amplifying the two targeted genes (nor-1 and ver-1) as these genes are considered as indicators of aflatoxin production, so this percentage indicates the possible contamination of aflatoxins in rice grains. Molecular method is proved as rapid and accurate detection system to differentiate the aflatoxigenic and non-aflatoxigenic isolates. Further studies on sequencing and the quantification of aflatoxins in rice grains and their relationship with the level of contaminating Aspergillus species are in progress.
Acknowledgements
The authors are grateful to African Union for the provision of funds through Pan African University Institute for Basic Sciences, Technology and Innovation (PAUSTI) to carry out this research.
References
- Haitham SA, Ali AB, Kamel AA, Khalid SE, Abdallah ME. Detection of Aspergillus and Penicillium species producing aflatoxin in rice grains imported into Saudi Arabia, 2013.
- Reiter E, Vouk F, Bohm J, Razzazi-Fazeli E. A?atoxins in rice a limited survey of products marketed in Austria. Food Control. 2010;21:988-91.
- Lai X, He Z, Liu R, Liu C. Potential for a?atoxin B1 and B2 production by Aspergillus ?avus strains isolated from rice samples. Saudi J Biol Sci. 2015;22:176-80.
- Sohaib A, Ali SW, Ahmed A, Mahmood R. Molecular characterization of fungal species isolated from rice grains. Institute of Agricultural Sciences, University of the Punjab, Quid-i-Azam Campus, Lahore, Pakistan, 2019.
- Schmidt M, Zannini E, Arendt EK. Recent advances in physical post-harvest treatments for shelf-life extension of cereal crops, 2018.
- Frisvad JC, Hubka V, Ezekiel CN, Hong SB, Novakova A. Taxonomy of Aspergillus section flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies Mycol. 2019;93:1-63.
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbio Mol Bio Rev. 2002;66:447-59.
- Lereau M, Gouas D, Villar S, Besaratinia A, Hautefeuille A. Interactions between hepatitis B virus and aflatoxin B1 Effects on p53 induction in Hepa RG cells. J Gen Virol. 2012;93:640-50.
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, World Health Organization, International Agency for Research on Cancer. Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. World Health Organization. 2002;82:171-300.
- Gemeda N, Woldeamanuel Y, Asrat D, Debela A, Lemma H. Assessment of aflatoxigeinic Aspergillus species in food commodities from local market of Addis Ababa. Research. 2014;1:1195.
- Lee J, Her JY, Lee KG. Reduction of aflatoxins (B1, B2, G1, and G2) in soybean-based model systems. Food Chem. 2015;189:45-51.
- Ruadrew S, Craft J, Aidoo K. Occurrence of toxigenic Aspergillus spp. and aflatoxins in selected food commodities of Asian origin sourced in the West of Scotland. Food Chem Toxicol. 2013;55:653-8.
- Naseer R, Sultana B, Khan M, Naseer D, Nigam P. Utilization of waste fruit-peels to inhibit aflatoxins synthesis by Aspergillus flavus: A biotreatment of rice for safer storage. Biores Technol. 2014;172:423-8.
- Tola M, Bedaso K. Occurrence, importance and control of mycotoxins: A review. Cogent Food Agri. 31;2:1191103.
- COMMISSION REGULATION (EC) No 401/2006 of 23 February 2006: Laying down the methods of sampling and analysis for the official control of the levels of mycotoxins in foodstuffs.
- Botton B, Breton A, Fèvre M, Gauthier S, Guy P. Moisissures utiles et nuisibles, importance industrielle, Masson, Paris,1990;p:349.
- Atlas RM. Handbook of Microbiological Media. CRC Press, Boca Raton, 2010.
- Pitt JI, Hocking AD. Fungi and Food Spoilage. New York: Springer Science, 3rd edition 2009.
- Varga J, Frisvad JC, Samson RA. Two new a?atoxin producing species and an overview of Aspergillus section. Flavi Stud Mycol. 2011;69:57-80.
- Mathur SA, Matur SB, Neergaard P. Detection of seed borne fungi in sorghum and location of Fusarium moniliforme in seed. Seed Sci Technol. 1975;3:683-90.
- Baqur MS, Shuhaib A, Albakri AH, Alwan SH, Almandil NB, et al. Optimal PCR Primers for Rapid and Accurate Detection of Aspergillus ?avus isolates, 2018.
- Raper KB, Fennell DI. The genus Aspergillus. The Williams & Wilkins Company, USA, 1765; pp: 370-76.
- White TJ, Bruns T, Lee SJ, Taylor JL. Ampli?cation and direct sequencing of fungal ribosomal RNA genes for phylogenetics, In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: A Guide to Methods and Applications, Academic Press, Inc, San Diego, CA USA, 1990; pp:315-22.
- Magan N, Hope R, Cairns V, Aldred D. Post-harvest fungal ecology: Impact of fungal growth and mycotoxin accumulation in stored grain. Eur J Plant Pathol 2003;109:723-30.
- Hina S, Ahmad AS, Shinawar WA. Characterization of fungal microbiota on rice grains from local markets of Lahore, 2014;37:35-40.
- Renu K, Agarwal MK, Bhagayavant SS, Verma P, Nagar DP. Detection of Aspergillus flavus using PCR method from fungus infested food grains collected from local market, 2018.
- Fariha I, Jalal IH, Khan AB, Asghar MA, Iqbal J. Prevalence of Aflatoxigenic Aspergillus in Food and Feed Samples, 2016.
- Reddy KRN, Reddy CS, Mangala UN, Muralidharan K. Site of Infection of Aspergillus sp. in seeds of rice cultivars. J Mycol 2006;36:271-7.
- Aydin A, Aksu H, Gunsen U. Mycotoxin levels and incidence of mould in Turkish rice. Environ Monitor Assess. 2010;10:1661-88.
- Majid Z, Maryam E. Molecular variation analysis of Aspergillus flavus using polymerase chain reaction-restriction fragment length polymorphism of the internal transcribed spacer rDNA region, 2016.
- Hadi AA, Carter D, Magan N. Discrimination between aflatoxigenic and non-aflatoxigenic Aspergillus section Flavi strains from Egyptian peanuts using molecular and 2011.
- Rodrigues P, Soares C, Kozakiewicz Z, Paterson RRM, Lima N. Identification and characterization of Aspergillus flavus and aflatoxins. In: Méndez-Villas A. (Editor) -Communicating current research and educational topics and trends in applied microbiology. Formatex. 2007;527-34.
- Rashid M, Khalil S, Ayub N, Ahmed W, Khan AG. Categorization of Aspergillus flavus and Aspergillus parasiticus isolates of stored wheat grains into aflatoxinogenics and non-aflatoxinogenics. Pak J Bot. 2009; 40: 2177-192.
- Degola F, Berni E, Dall'Asta C, Spotti E, Marchelli R, et al. A multiplex RT-PCR Approach to Detect Aflatoxigenic Strains of Aspergillus flavus. J Appl Microbiol. 2007;103:409-17.
- Houshyarfard M, Rouhani H, Falahati-Rastegar M, Malekzadeh-Shafaroudi S, Mehdikhani Moghaddam E. Gene Deletion Patterns in non-aflatoxigenic Strains of Aspergillus flavus. Mycologia Iranica. 2014;2:87-97.
- Khattab A, Shekhany M, Rostam RK. Detection of A?atoxigenic Aspergillus Flavus in maize grains and soils in sulaimani province using molecular approaches. Journal of Zankoy Sulaimani. 2016;18?.
- Hussain A, Afzal A, Irfan M, Malik KM. Molecular detection of aflatoxin producing strains of Aspergillus flavus from Peanut (Arachis Hypogaea). Plant Pathology Journal. 2006; 5: 115-124.
Author Info
Youmma Douksouna1*, Andrew Nyerere1,2, Joel Masanga3, Steven Runo3 and Zachée Ambang42Department of Medical Microbiology, Jomo Kenyatta University of Agriculture and Technology, Nairobi, Kenya
3Department of Biochemistry and Biotechnology, Kenyatta University, Nairobi, Kenya
4Department of Plant Biology and Biotechnology, University of Yaounde, Cameroon
Citation: Douksouna Y, Nyerere A, Masanga J, Runo S, Ambang Z (2019) Towards Managing and Controlling Aflatoxin Producers within Aspergillus Species in Infested Rice Grains Collected from Local Market In Kenya. Appli Microbiol. Open Access. 5:161 doi: 10.35248/2471-9315.19.5.160
Received: 19-Jul-2019 Accepted: 30-Jul-2019 Published: 07-Aug-2019
Copyright: © 2019 Douksouna Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.