Research Article - (2024)Volume 9, Issue 3
Gastrointestinal Stromal Tumors (GIST) has the potential for malignant transformation during recurrence and distant metastasis, but the mechanism of metastasis and related gene targets are unknown. In this study, a bioinformatics approach was used to identify potential therapeutic targets for GIST transfer inhibition. Firstly, 761 differentially expressed genes were identified based on the GSE136755 dataset and GSE21315 dataset. Enrichment analysis, protein-protein interaction network, and key gene identification were carried out successively. In addition, tissue specific expression analysis and prediction model construction of key genes were carried out. The results showed that the tissue-specific expression of five key genes (ALB, VEGFA, CDH1, JUN, CXCL8) increased significantly, and the prediction model constructed had a good prediction effect. In conclusion, the identified key genes (ALB, VEGFA, CDH1, JUN, CXCL8) may be used as therapeutic targets to inhibit the malignant progression of GIST.
GIST; Distal transfer; Targeted genes; Bioinformatics analysis; Diagnostic model
Gastrointestinal Stromal Tumors (GIST) is the most common mesenchymal tumors of the gastrointestinal tract and is generally defined as KIT (CD117) positive tumors with specific histologic features [1-4]. GIST has abdominal recurrence and liver metastases and has the potential for malignant transformation [5-7]. The exact pathogenesis of GIST remains to be fully elucidated, and there is no cure. Current GIST treatments mainly include open surgery, laparoscopic surgery, endoscopic therapy, and drug targeted therapy, but the treatment effect is far less than expected [8-10]. The postoperative recurrence and metastasis rate of high risk patients with GIST treated by surgical resection can reach 55%~90% [11]. The 5 years survival rate of patients without metastases who have completely resected the primary lesion is 50%~65%, the 5 years survival rate of those who cannot be completely resected or metastasized is less than 35%, and the overall survival of those who cannot be resected is 9 months~12 months [12-15].
Current studies of early lesions of GIST have found that most GISTs produce functional mutations in the two receptor tyrosine kinase genes KIT (60%-85%) and PDGFRA (5%-10%), resulting in conformational changes in their respective proteins that activate downstream signaling pathways, including RAS/RAF/MAPK and PI3K/AKT/mTOR. However, the study of potential target genes related to immunity and pathway during the progression of GIST distal metastasis is weak, and there is a lack of in-depth excavation analysis.
This study aims to explore the hub genes and potential mechanisms of GIST transfer using bioinformatics methods. Raw data from GIST transfer samples and primary control samples were downloaded from the GEO database. Through enrichment analysis and Protein–Protein Interaction (PPI) network analysis, key immune-related genes in the progression of GIST distal metastasis were identified, and potential targets for distal GIST metastasis progression were identified, so as to develop effective treatments to contain GIST distal metastasis and disease progression.
Data sources
Our datasetsGSE136755 and GSE21315 are both from the GEO database and use the normalize quantiles function in the R software preprocess core package for data normalization. Convert probe IDs to gene symbols, and remove probes for multiple genes. The remove Batch Effect function in the R software Limma software package was used to remove the batch effect, and the data standardization was evaluated by boxplot for the data preprocessing results, and the data batch effect was evaluated by comparing the visual PCA plot before and after batch removal.
Bioinformatics analysis
Differential expression of mRNA was investigated by the limma software package of R software. Adjusted P values were analyzed in GEO to correct for false-positive results. Gene function enrichment analysis uses the GO annotation of genes in the R package org.Hs.eg.db. The latest KEGG Pathway genes were obtained from the KEGG rest API and enriched using the R package clusterProfiler (version 3.14.3) to obtain gene set enrichment results. Set the minimum gene set to 5, the maximum gene set to 5000, and P value of <0.05 and a FDR of <0.25 were considered statistically significant.
The gene co-expression network was constructed by using the "WGCNA" package in R software, and the gene set was identified based on the weighted correlation by weighted expression correlation, hierarchical clustering analysis was carried out, and different gene modules were obtained according to the set standard segmentation clustering results, which were represented by branches and different colors of the cluster tree. Build a PPI network through the STRING database.
The confidence score ≥ the 0.4 criteria for screening. Use the Cytoscape v3.7.2 and CytoHubba plugin (version 0.1) sieve to visualize and identify PPI networks. The genes of Top 20 were screened out by the screening algorithm (affinity) for follow-up research. Information on immune related genes is available through the ImmPort Portal. Key genes from this study were obtained from immune related genes and Hub genes using Wayne diagrams. Wilcox tests were used to analyze the differences in the expression of key genes in primary GIST and metastatic GIST tissues. GSEA software (version 3.0) was obtained from the GSE website to divide samples into high expression groups (≥ 50%) and low expression groups (<50%) based on the expression level of key genes. The c2.cp.kegg.v7.4. symbols.gmt subset was downloaded from the molecular signatures database to evaluate the pathways and molecular mechanisms, and the final result was presented in the "enrich plot" package in the R software.
Finally, a predictive model is constructed based on the logistic regression algorithm of R software package.
Statistical analysis
The data used in this study are all standardized TPM (Transcripts Per kilobase of exon model per million mapped reads) data, and their data distribution is close to normal distribution, so both the T-test and the rank sum test can be used to evaluate the difference between the two groups. In this study, a p value of less than 0.05 was considered statistically significant.
A total of 62 patients with primary GIST and 11 patients with metastatic GIST were included in this study. 761 tissue differentially expressed genes were identified by the GSE136755 and GSE21315 datasets. The DEGs were successively analyzed by functional enrichment and pathway enrichment, and a PPI network was constructed, and the top 20 Hub genes were screened. Acquire immunologically relevant genes through the ImmPort Portal and build Venn graphs with 20 Hub genes, mining key genes in GIST remote transfer. Enrichment analysis of key genes was performed by GSEA software and c2.cp.kegg.v7.4.symbols.gmt subsets to assess relevant pathways and molecular mechanisms. Finally, a predictive model is constructed based on logistic regression algorithm to analyze the prognostic correlation of key genes in order to develop effective treatments to curb the progress of GIST distal metastasis (Figure 1).
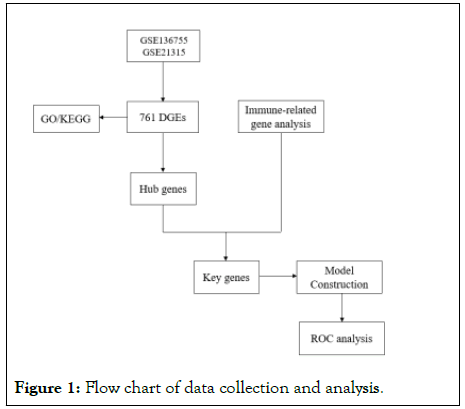
Figure 1: Flow chart of data collection and analysis.
Differential expression analysis
We used the R software Limma package to analyze the expression differences between GSE13675 dataset and GSE21315 dataset in early GIST and metastatic GIST tissues, and screened out significantly different genes. After setting the cut-off values P < 0.05 and |log2(FC)|>1, a total of 761 DEGs were identified (Figure 2).
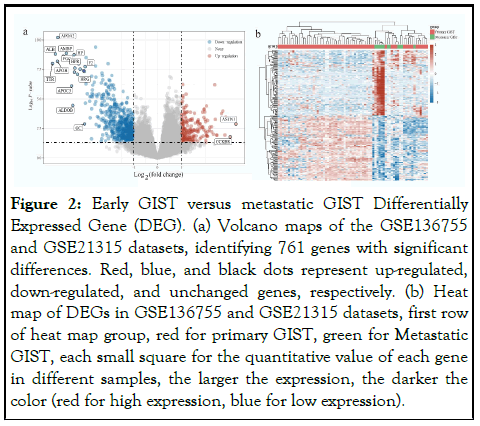
Figure 2: Early GIST versus metastatic GIST Differentially Expressed Gene (DEG). (a) Volcano maps of the GSE136755 and GSE21315 datasets, identifying 761 genes with significant differences. Red, blue, and black dots represent up-regulated, down-regulated, and unchanged genes, respectively. (b) Heat map of DEGs in GSE136755 and GSE21315 datasets, first row of heat map group, red for primary GIST, green for Metastatic GIST, each small square for the quantitative value of each gene in different samples, the larger the expression, the darker the color (red for high expression, blue for low expression).
Gene set functional enrichment analysis
We performed gene set function enrichment analysis on the screened DEGs to determine their relevant pathways or biological processes in Primary GIST and Metastatic GIST. Using the gene GO annotations in the R package org.Hs.eg.db, GO annotations are divided into three categories: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The DEGs of the BP group were mainly enriched in the regulation of biological quality, small molecule metabolic process and lipid metabolic process. Changes in these biological processes are critical for the progression of distant metastasis in GIST patients. The DEGs of the CC group were mainly enriched in the extracellular region, vesicle, extracellular vesicles and extracellular organelles. DEGs in the MF group were mainly enriched in signaling receptor binding, suggesting that DEGs may play a key role in regulating signaling receptor binding (Figure 3a).
Genes for the latest KEGG pathway were obtained from the KEGG rest API, KEGG analysis of DEGs using the R package ClusterProfiler (version 3.14.3) and found that they were mainly rich in "metabolic pathways","PI3K-Akt" signaling pathway" and "complement and coagulation cascades" (Figure 3b). Alterations in metabolic pathways are one of the characteristics of tumor cells, PI3K-Akt signaling is associated with tumor suppression, imbalance of complement and coagulation cascades and increased release of certain coagulation factors play a vital role in promoting hypercoagulable states and vascular endothelial dysfunction. These three pathways are closely related to human cancer, providing multiple potential targets for cancer treatment. Based on enrichment analysis, we further performed WGCNA analysis on the differential genes, constructed a gene coexpression network using the "WGCNA" package in R software, expressed gene correlations with weights, segmented clustering results, and represented different gene modules by branching and different colors of the cluster tree (Figure 4).
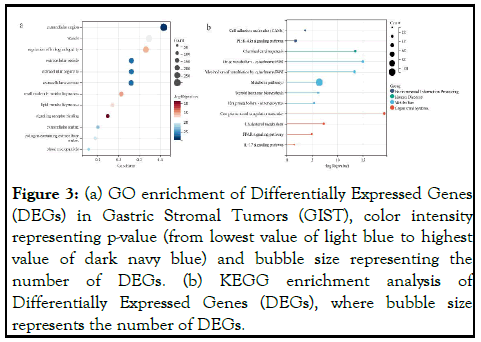
Figure 3: (a) GO enrichment of Differentially Expressed Genes (DEGs) in Gastric Stromal Tumors (GIST), color intensity representing p-value (from lowest value of light blue to highest value of dark navy blue) and bubble size representing the number of DEGs. (b) KEGG enrichment analysis of Differentially Expressed Genes (DEGs), where bubble size represents the number of DEGs.
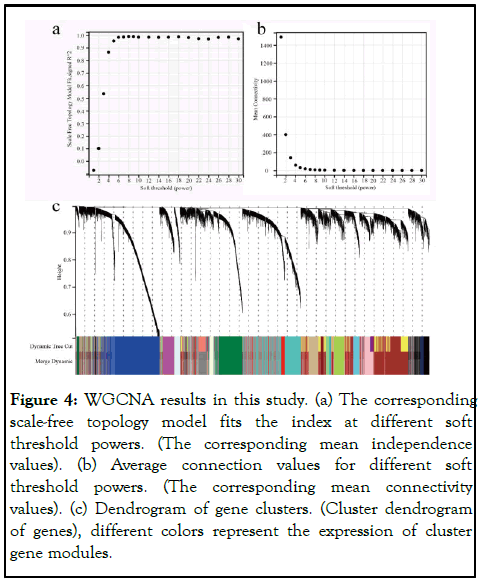
Figure 4: WGCNA results in this study. (a) The corresponding scale-free topology model fits the index at different soft threshold powers. (The corresponding mean independence values). (b) Average connection values for different soft threshold powers. (The corresponding mean connectivity values). (c) Dendrogram of gene clusters. (Cluster dendrogram of genes), different colors represent the expression of cluster gene modules.
PPI network construction and key gene identification
In this study, a PPI network was constructed from the STRING database (Figure 5). The confidence score ≥ the 0.4 criteria for screening. Use cytoscape v3.7.2 and CytoHubba plugin (version 0.1) to filter out hub genes with Top20 scores, including ALB, FN1, APOB, VEGFA, AHSG, APOA1, F2, AFP, SERPINC1, FGG, TTR, SERPINA1, CDH1, JUN, CXCL8, MYC, PLG, APOC3, CPB2, FGB.
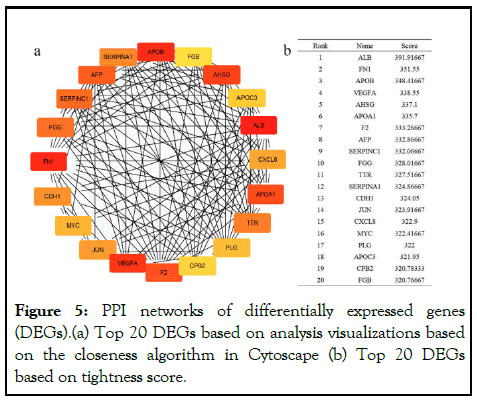
Figure 5: PPI networks of differentially expressed genes (DEGs).(a) Top 20 DEGs based on analysis visualizations based on the closeness algorithm in Cytoscape (b) Top 20 DEGs based on tightness score.
1793 immunologically relevant genes and 20 hub genes were obtained through the ImmPort portal to construct a Venn map and 5 key genes were identified, namely ALB, VEGFA, CDH1, JUN, CXCL8 (Figure 6). These five key genes not only have significant expression differences, but also are related to biological overflows such as immunity, biological mass regulation, small molecule metabolism and lipid metabolism.
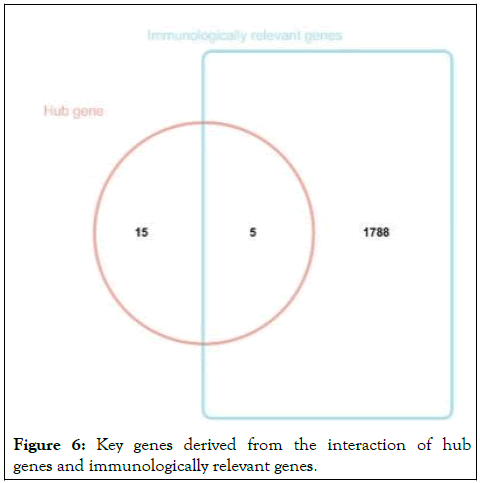
Figure 6: Key genes derived from the interaction of hub genes and immunologically relevant genes.
Differential expression analysis of key genes
Wilcox tests were used to analyze the differences in the expression of 5 key genes in primary GIST and metastatic GIST. The ALB, VEGFA, CDH1, JUN, and CXCL8 genes were significantly lower in early GIST and significantly higher in metastatic GIST, indicating that there are differences in the biological processes and molecular mechanisms of key genes in GIST at different times (Figure 7).
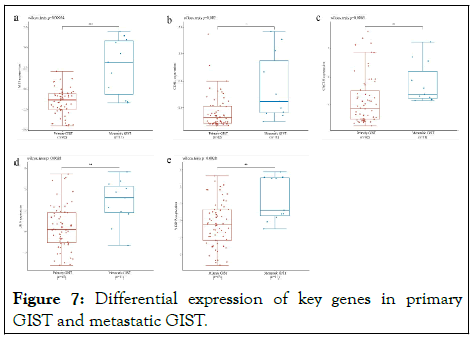
Figure 7: Differential expression of key genes in primary GIST and metastatic GIST.
GSEA enrichment analysis by key gene
To further explore the influence of the biological functions of these five key genes on GIST, we performed enrichment analyses on them. Samples were divided into high expression groups (≥ 50%) and low expression groups (<50%) by GSEA software (version 3.0) and from the molecular.
Signatures database the c2.cp.kegg.v7.4. symbols.GMT subset was downloaded to evaluate related pathways and molecular mechanisms. The final result is presented using the "enrich plot" package in R software.
GSEA enrichment analysis by Key genes revealed that they are associated with biological processes (progesterone-mediated oocyte maturation, glycolytic gluconeogenesis, galactose metabolism, starch and sucrose metabolism, galactose metabolism) and multiple pathways (VEGF signaling pathway, B-cell receptor signaling pathway, complement and coagulation cascade) in multiple cancers (thyroid cancer, endometrial cancer, colorectal cancer, kidney cell and cancer bladder cancer) (Figure 8). These findings validate the key role of these five key genes in GIST and provide new information for the study of potential targets for other cancers.
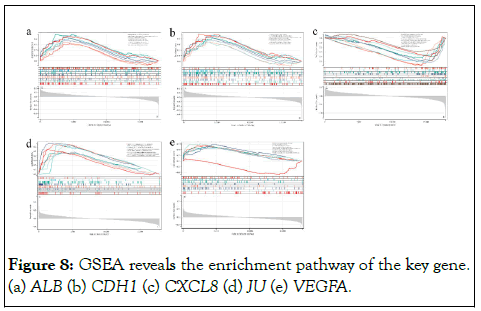
Figure 8: GSEA reveals the enrichment pathway of the key gene. (a) ALB (b) CDH1 (c) CXCL8 (d) JU (e) VEGFA.
Construction of diagnostic models
Finally, in order to confirm the prognosis prediction of GIST of these five key genes, we construct five key gene prediction models based on the logistic regression algorithm. Loss regression analysis showed that these five gene prediction models had good diagnostic performance, with an AUC of 0.9 and the model was reliable (Figure 9).
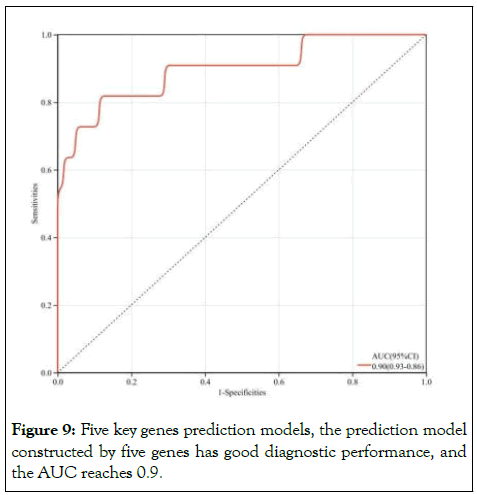
Figure 9: Five key genes prediction models, the prediction model constructed by five genes has good diagnostic performance, and the AUC reaches 0.9.
GIST is the most common mesenchymal tumor of the digestive tract, and its incidence is increasing at an alarming rate worldwide. GIST is subtle and is often found incidentally during other procedures [16-19]. GIST has the dual characteristics of benign and malignant and cannot be simply judged as benign or malignant and needs to be evaluated comprehensively based on a variety of factors [20]. GIST may seem benign in the short term, but over time, almost all GITs will have malignant behaviors, such as increasing size, metastasis, and postoperative recurrence. Thus, GIST can be considered an aggressive tumor with malignant potential, which is determined by tumor size, location, mitotic index, and pathological results. The higher the malignant potential, the greater the potential for death, so it is important to diagnose and treat distal metastatic progression of GIST as early as possible.
To further explore key target genes in GIST distal transfer progression, 761 DEGs were obtained from selected datasets GSE136755 and GSE21315. Through GO and KEGG enrichment analysis, it was found that these genes are mainly involved in signaling receptor binding, regulation of biological quality, small molecule metabolic process and lipid metabolic process. It is enriched in metabolic pathways and PI3K-Akt signaling pathways. These biological processes and pathways play a very important role in the functioning of the body and the development of tumors. Regulation of biological quality is an important way for the body to maintain homeostasis in the internal environment. The metabolome produced by the small molecule metabolic process is often used as a biomarker for diagnosing or predicting disease, and can also bind directly to proteins to regulate protein function, modify disease related signaling pathways, and influence disease progression.
PI3K-Akt Phosphatidylinositol 3 Kinase (PI3Ks) signaling involves the regulation of various cellular functions such as proliferation, differentiation, apoptosis, and glucose transport. It can be seen that these differentially expressed genes are inseparable from the development and change of GIST distal transfer, and are crucial for the exploration of therapeutic targets of GIST.
We further screened out the hub genes with synergistic correlation through weighted co-expression and PPI networks, and obtained 5 key genes, including ALB, VEGFA, CDH1, JUN and CXCL8, by constructing Venn diagrams with immune related genes. Through comparison, it can be found that the expression of these five key genes in the process of GIST distal transfer is significantly increased, which has an important impact on multiple biological processes or immune changes in GIST distal metastasis progression. An association between highly expressed VEGFA in GIST and poor prognosis in GIST patients has been shown in previous GIST studies, and CXCL8 has multiple roles in the Tumor Microenvironment (TME) and may be a potential target for cancer therapy, which was confirmed in our study. Key genes such as ALB, CDH1, and JUN have also been studied in other diseases. ALB is produced by hepatocytes, maintains intravascular pressure and balances blood pH, and is strongly associated with nutritional status and systemic inflammation. In cancer progression, ALB expression is inhibited by cytokines and growth factors produced by tumor cells and immune cells. In addition, ALB has antioxidant functions and anticancer effects. JUN is a major component of the transcription factor complex AP-1, which is involved in basic cellular processes and controls cellular stimulus responses to regulate proliferation, differentiation, oncogenic transformation, and apoptosis. CDH1 inhibits cell proliferation and HNSCC invasion and has the function of tumor suppressor genes in head and neck cancer. CXCL8 is one of the most important proinflammatory factors and plays a crucial role in many inflammatory diseases, including UC. In some cancers, VEGFA transcription and tumor angiogenesis are inhibited by targeting transcriptional regulators NCOR2 and SerRS. Our research further sheds light on their important diagnostic and therapeutic role as target genes in GIST. In the future, we can further explore other biological processes and molecular mechanisms of these five key genes in GIST tissues. Our study also had several limitations; first, our gene set was constructed and validated through retrospective data from public databases, requiring more forward looking, and real world data to validate its clinical utility. Secondly, the molecular regulatory mechanism of GIST related genes in immunity should be further studied on a large scale to explore other functions and values of these key genes.
In conclusion, the immune related genes of ALB, VEGFA, CDH1, JUN and CXCL8 mined in this study are closely related to the occurrence and development of GIST distal transfer progression, and the predicted model constructed has good diagnostic performance. Combined with the analysis of relevant biological processes in our study, it can be seen that these immune related genes and multiple pathways may become new targets for GIST therapy, and each immune related key gene can be independently analyzed in the future in order to develop more effective cancer treatment methods and provide new directions for subsequent targeted drug development. Large scale, multicenter in-depth studies are needed to further validate these findings in future studies.
Health innovation special project of Hebei provincial key R and D program (223777101d).
DC, RF: Methodology, data curation, writing and validation. ML: Methodology and validation. WG: Reviewing, editing and funding acquisition. JF: Conceptualization, supervision and funding acquisition.
None.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Cui D, Li M, Fu R, Guo W, Fei J (2024) Potential Therapeutic Targets for Distant Metastasis of Gastrointestinal Stromal Tumors: Bioinformatics Analysis. Immunogenet Open Access. 9:222.
Received: 06-May-2023, Manuscript No. IGOA-23-23932; Editor assigned: 09-May-2023, Pre QC No. IGOA-23-23932 (PQ); Reviewed: 23-May-2023, QC No. IGOA-23-23932; Revised: 26-Jul-2023, Manuscript No. IGOA-23-23932 (R); Published: 26-Mar-2024 , DOI: 10.35248/IGOA.24.9.227
Copyright: © 2024 Cui D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.