
Journal of Clinical Chemistry and Laboratory Medicine
Open Access
ISSN: 2736-6588

ISSN: 2736-6588
Research Article - (2023)Volume 6, Issue 4
Background: Reporting directly measured homogeneous Direct Low-Density Lipoprotein Cholesterol (D-LDL) becomes imperative especially in tertiary level laboratories which have to frequently deal with dyslipidaemic specimens. In absence of credible studies involving dyslipidaemic specimens on comparability of test results between different platforms or between different platforms and reference methods, reporting of D-LDL becomes very uncertain.
Methods: The current study includes 328 subjects classified from Type I to Type V according to Fredrickson classification of dyslipidaemia. Standard lipid profile including D-LDL was tested on their serum specimens and D-LDL was repeat tested after saline dilution on three platforms viz AU5800, Alinity ci and Cobas Pure. Calculated LDL-Cholesterol for all the specimens were derived from the NIH equation proposed by Sampson, et al.
Results: Mean Absolute Percent Variation (MAPV) between D-LDL and C-LDL for each class interval was found to increase with increasing triglycerides concentrations of the specimen and at the two extremes of non-High-Density Lipoproteins (HDL) cholesterol concentrations. Passing-Bablok regression, Bland-Altman plot and Receiver Operating Characteristic curves constructed for each dyslipidaemia phenotype revealed that AU5800 outperformed the other two for Type II and III specimens, while Alinity ci and Cobas Pure outperformed AU5800 for Types I, IV and V specimens.
Conclusion: Variation in test results of D-LDL in dyslipidaemic specimens on most widely used platforms is a matter of concern as it might lead to misclassifications in diagnosis and treatment monitoring.
Direct LDL; Homogeneous LDL; LDL comparison; ROC curves
To make reporting cost effective, most clinical laboratories now- a-days resort to reporting lipid profile with the LDL-Cholesterol (LDL-C) as a calculated parameter or not to report it altogether in case of lipaemic specimens. As a result, much emphasis has been laid on developing a foolproof equation for calculating LDL-C, including validation against ultracentrifugation, validation against β-quantification and even machine learning [1-3]. On the other hand, demand for directly measured homogeneous assays for LDL-C is on the rise, mainly in tertiary and quaternary level reference laboratories, which have to frequently deal with dyslipidaemic samples referred from the lower levels. When it comes to directly measured LDL-C (D-LDL) methods, there have been comparison studies which agree to their comparability, but there is a dearth of information regarding their comparability while dealing with dyslipidaemic specimens [4].
It is in this area that the present study envisages adding some useful information. Three popular homogeneous LDL-C methods were taken up-dedicated LDL-C reagents of AU5800 (Beckman- Coulter), Alinity ci (Abbott Laboratories) and Cobas Pure (Roche Diagnostics). Three hundred and twenty-eight dyslipidaemic subjects were selected, classified from Type I through Type V according to Fredrickson-Levy-Lees phenotypic classification and their standard lipid profiles estimated [5]. In the absence of any access to a β-quantification platform, the measured D-LDL results were compared to the equation proposed by Sampson, et al. hereafter referred to as C-LDL, which was itself validated against β-quantification [2]. The other reason for choosing C-LDL is that the original study was also based on dyslipidaemic specimens, which matches the aim of the current study. Though Sampson et al excluded Type III samples from their study, at least one study is available, which validates the veracity of their formula even in Type III subjects. Hence Type III subjects were included in the present study [6]. The other option available was to compare with a formula validated against ultracentrifugation, but since this study includes a substantial number of lipaemic specimens, it was decided against using the corresponding equation, as ultracentrifugation is known to underestimate Very-Low-Density Lipoprotein Cholesterol (VLDL-C) levels in hypertriglyceridaemic specimens [1,7].
This cross-sectional non-interventional observational study was undertaken at Drs. Tribedi & Roy Diagnostic Laboratory, 93 Park Street, Kolkata, India, which is a private undertaking tertiary-level referral laboratory, from November 2022 to July 2023. Informed consent from the study subjects and ethical clearance from the institutional ethical committee was duly undertaken for this study. Overnight fasting dyslipidaemic results were segregated from the routine laboratory workflow, the corresponding subjects were identified and contacted for an interview. The subjects who responded to a standard questionnaire underwent a brief clinical examination before finally being classified into their corresponding dyslipidaemia phenotype. Those undergoing lipid lowering therapy or having active liver and/or kidney diseases were excluded from the study. Demographic characteristics of the subjects are enumerated in Table 1.
| Type I | Type II | Type III | Type IV | Type V | Total | ||
|---|---|---|---|---|---|---|---|
| N | 16 | 42 | 61 | 126 | 83 | 328 | |
| Median age (Years) | 23 | 37 | 26 | 42 | 48 | 43 | |
| Male: Female ratio | 0.95 | 1.57 | 2.15 | 1.07 | 0.94 | 1.34 | |
| Total cholesterol | Range | 53-521 | 132-1115 | 125-715 | 57-335 | 133-524 | 53-1115 |
| Median | 168 | 395 | 332 | 194 | 281 | 262 | |
| Triglycerides (mg/dL) | Range | 468-5060 | 63-386 | 100-886 | 194-694 | 700-3404 | 63-5060 |
| Median | 1115 | 176 | 336 | 569 | 964 | 571 | |
| Non-HDL cholesterol (mg/dL) | Range | 53-521 | 63-1017 | 83-647 | 48-297 | 113-507 | 48-1017 |
| Median | 140 | 332 | 274 | 158 | 245 | 215 | |
| D-LDL (AU5800), in mg/dL | Range | 0-153 | 70-657 | 68-417 | 46-216 | 65-371 | 0-657 |
| Median | 66 | 276 | 223 | 122 | 149 | 148 | |
| D-LDL (Alinity ci), in mg/dL | Range | 17-105 | 61-797 | 47-470 | 10-183 | 28-295 | 10-797 |
| Median | 45 | 289 | 208 | 81 | 79 | 100 | |
| D-LDL (Cobas Pure), in mg/dL | Range | 14-79 | 65-950 | 21-507 | 11-197 | 22-174 | 11-950 |
| Median | 29 | 297 | 221 | 80 | 81 | 100 | |
| C-LDL (mg/dL) | Range | -17-348 | 50-846 | 52-429 | 17-165 | 16-196 | -17-846 |
| Median | 20 | 292 | 206 | 67 | 70 | 89 | |
Note: The Types denote dyslipidaemia phenotypes as described by Fredrickson, et al. D-LDL: Directly measured LDL-Cholesterol, C-LDL: LDL-Cholesterol calculated by the NIH equation proposed by Sampson, et al. To convert Total cholesterol, LDL-Cholesterol and non-HDL-Cholesterol to millimoles per litre, multiply by 0.0259; to convert Triglycerides to millimoles per litre, multiply by 0.0113.
Table 1: Demographics of the study population.
After an initial measurement on the day of collection, the serum specimens were aliquoted and stored at -20°C. After the response was obtained from the subject, preferably within a week, the corresponding serum specimen was thawed, diluted with 0.9% NaCl and its LDL-C was retested in an effort to negate the effect of any interferant present in the original specimen. D-LDL was measured from the undiluted and the diluted specimens on the three platforms viz AU5800, Alinity ci and Cobas Pure. Performance specifications of the D-LDL reagents are elaborated in Table 2. C-LDL was determined for all the subjects. Data was segregated into five broad groups according to the type of dyslipidaemia present viz Type I, Type II, Type III, Type IV and Type V (Table 1).
| AU5800 | Alinity ci | Cobas pure | |
|---|---|---|---|
| Method | Homogeneous, selective chemoprotection, colorimetric. | Homogeneous, selective detergent solubility, colorimetric. | Homogeneous, selective micellary solubilization, colorimetric. |
| Traceability | US CDC reference method | US CDC Abell-Kendall reference method | β-quantification method |
| Linearity | 10-400 mg/dL | 1-800 mg/dL | 3.87-549 mg/dL |
| Precision | 2.34-2.71 % CV | 1.5-1.6 % CV | 1.6-1.7 % CV |
| Accuracy w.r.t. established method | Correlation coefficient, r=0.966 | Correlation Coefficient, r=0.96 | Correlation coefficient, r=0.992 |
| Lipaemic interference | <10% up to 1000 mg/dL of TGs | Negligible up to 1293 mg/dL of TGs | Negligible up to L index of 1000 |
| Limitations | +30% bias against ref. method in Type III subjects; Waldenström’s macroglobulinaemia. | - | Abnormal results in liver diseases, Waldenström’s macroglobulinaemia. |
Note: All data as per claims of the respective manufacturers. To convert LDL-Cholesterol to millimoles per litre, multiply by 0.0259; to convert Triglycerides to millimoles per litre, multiply by 0.0113.
Table 2: Assay performance characteristics of D-LDL reagents.
Data analysis was done on Excel worksheets (Microsoft Corp) and R-Programme/ Shiny web application. The data subsets were first tested for normality. Since the sample sizes were small, typically ~50, the Lilliefors Test was used [8]. All the data subsets were proven to lack normality. Hence further statistical treatment of the data was done with methods which do not require normality of the data. Firstly, variation between C-LDL and D-LDL was analyzed with respect to the corresponding triglycerides and non-HDL results by segregating the entire data into six series: (i) AU5800, representing results of undiluted samples on AU5800; (ii) Alinity, representing results of undiluted samples on Alinity ci; (iii) Cobas, representing results of undiluted samples on Cobas Pure; (iv) AU Diluted, representing results of diluted samples on AU5800; (v) Alinity Diluted, representing results of diluted samples on Alinity ci and (vi) Cobas Diluted, representing results of diluted samples on Cobas Pure. Then the entire data was segregated into the five dyslipidaemia phenotypes: Type I, Type II, Type III, Type IV and Type V. C-LDL and D-LDL results of each type were analyzed by Passing-Bablok regression (PB regression), a method which is distribution-free i.e., it makes no assumption about the underlying data distribution [9,10]. Composite Bland-Altman plots (BA plots), again a method free from data distribution requirements, were constructed separately for the five types, comparing the D-LDL and C-LDL results, highlighting the six series of data described above. Finally, composite Receiver Operating Characteristic (ROC) curves were constructed separately for the five types based on the percentage variation of the D-LDL results from the corresponding C-LDL results and using 12% as the cut-off based on the National Cholesterol Education Program (NCEP) recommendations for Total Allowable Error Goal for D-LDL estimation [11-13]. It may be noted that ROC curves also do not require normality of the data.
The entire data set was segregated into class intervals according to their triglycerides results and alternately according to their non-HDL results and arranged into the six groups viz AU5800, Alinity, Cobas, AU Diluted, Alinity Diluted and Cobas Diluted. Mean Absolute Percent Variation (MAPV), defined as the average of the absolute values of percentage deviation between C-LDL and D-LDL results, were determined for each class interval. These MAPV values were finally plotted against their corresponding class intervals (Figure 1). As is evident from the figure, variation in LDL results increased with increasing Triglycerides (TG) levels in all the six series. However, results of AU5800 varied more in comparison to the other platforms. When analyzed with respect to the corresponding non-HDL results, variation of LDL results increased in both extremes of non-HDL values and AU5800 results varied more in comparison with the other platforms. One more observation worth noting is that dilution of the specimens did not produce any improvement: for AU5800, the variation worsened; for the other platforms, it remained the same. In fact, the results of the four series viz Alinity, Cobas, Alinity Diluted and Cobas Diluted mirrored each other. The six data series in each panel represent LDL-Cholesterol estimations on the undiluted specimens performed on the three platforms AU5800, Alinity ci and Cobas Pure; and those on the same specimens diluted with normal saline and retested on the same three platforms. To convert Triglycerides to millimoles per litre, multiply by 0.0113; to convert Non-HDL-Cholesterol to millimoles per litre, multiply by 0.0259 (Figure 1).
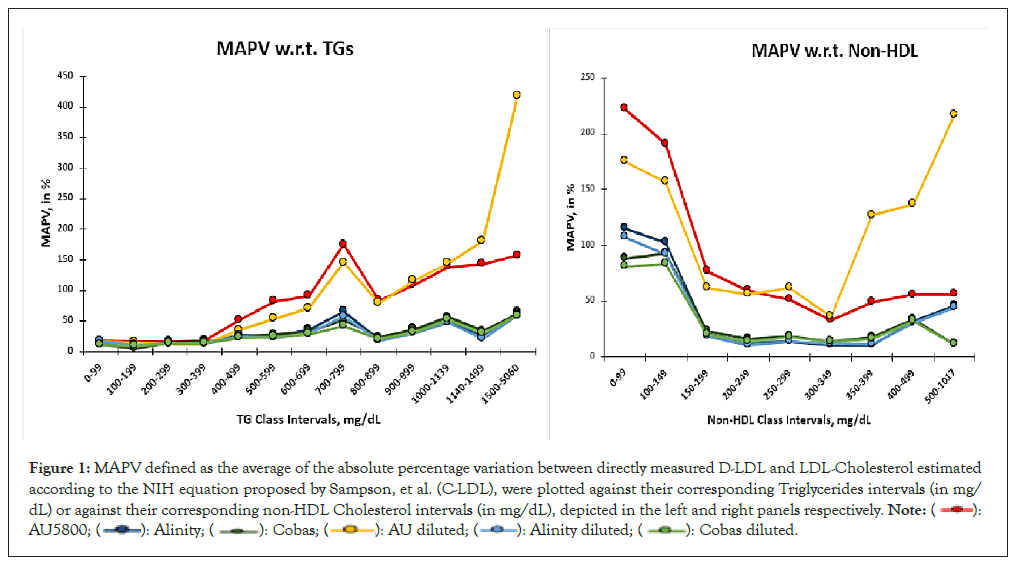
Figure 1: MAPV defined as the average of the absolute percentage variation between directly measured D-LDL and LDL-Cholesterol estimated
according to the NIH equation proposed by Sampson, et al. (C-LDL), were plotted against their corresponding Triglycerides intervals (in mg/
dL) or against their corresponding non-HDL Cholesterol intervals (in mg/dL), depicted in the left and right panels respectively. 
While it was evident that the variation in D-LDL results was dependent upon the corresponding TGs and Non-HDL-C concentrations, it was nevertheless imperative to determine if this variation was related to the dyslipidaemia phenotypes. Hence, in the next level of data analysis, the entire data set was segregated according to the dyslipidaemia phenotypes. In the study population (N=328), Type IV subjects were most numerous (N=126), followed by Type V (N=83), while Type I subjects were the most infrequent (N=16). Type II and III had 42 and 61 subjects respectively. In each of the Types, the relationship between D-LDL and C-LDL was examined by PB regression. Significant deviation from linear relationship was observed in Type I specimens, so much so, that PB regression could not be performed; instead, Deming regression was undertaken for the Type I results (Table 3). Though linear relationships were confirmed in the other types, significant deviation in slope and significantly large intercept were observed in the results of Type V specimens. Similar deviations, albeit in lesser degrees, were observed in the D-LDL results in Type IV specimens. In other words, presence of significant lipaemia affected D-LDL results in all the platforms. The best-fit regression lines were obtained for the Types II and III results. Due to the non-parametric nature of the data sets, Spearman’s correlation analysis was done for all the five types and their coefficients (ρ) determined (Table 3).
| Data Set | Regression model | Slope | Intercept | Spearman coefficient, ρ |
|---|---|---|---|---|
| Type I | Deming | 0.15 | 57.01 | 0.16 |
| Type II | Passing-bablok | 0.97 | -0.49 | 0.97 |
| Type III | Passing-bablok | 1.04 | -0.19 | 0.929 |
| Type IV | Passing-bablok | 1.15 | 6.29 | 0.806 |
| Type V | Passing-bablok | 1.73 | -22.5 | 0.538 |
Table 3: Regression results.
Regression analysis revealed that variation in D-LDL results indeed depended on the type of dyslipidaemia phenotype. It was, however, important to examine if any of the three platforms was more responsible than the others in contributing to this variation within each dyslipidaemia phenotype. Hence Bland-Altman Plots (BA plots) were constructed with (C-LDL+D-LDL)/2 as the abscissa and (C-LDL-D-LDL) as ordinate for the five dyslipidaemia phenotypes separately. The points were colour coded according to the six data series as described in the ‘Materials and Method’s section. The six data series in each panel represent LDL-Cholesterol estimations on the undiluted specimens performed on the three platforms AU5800, Alinity ci and Cobas Pure; and those on the same specimens diluted with normal saline and retested on the same three platforms. To convert LDL-Cholesterol to millimoles per litre, multiply by 0.0259. (Figure 2). In Type I specimens, the BA plot is characterized with very wide limits of agreement and yet ends up with significant number of outliers from all the three platforms, especially in the middle range of mean values. Since the sample size of Type I specimens is miniscule, any comparative analysis between the three platforms tested must be accepted with caution. The degree of agreement between the two data series in Type II specimens is much greater in comparison with Type I; the limits of agreement have narrowed down and outliers are mostly restricted to the higher extreme of mean values. BA plot of Type III specimens also depicts fair degree of agreement for all the platforms. When it comes to Type IV specimens, a definite bias is noted (-19 mg/dL); almost all the AU5800 results on undiluted and diluted specimens lie on one side of even the Bias line, indicating that the corresponding D-LDL results are significantly greater than their C-LDL estimates. No such trend is evident for the other two platforms.
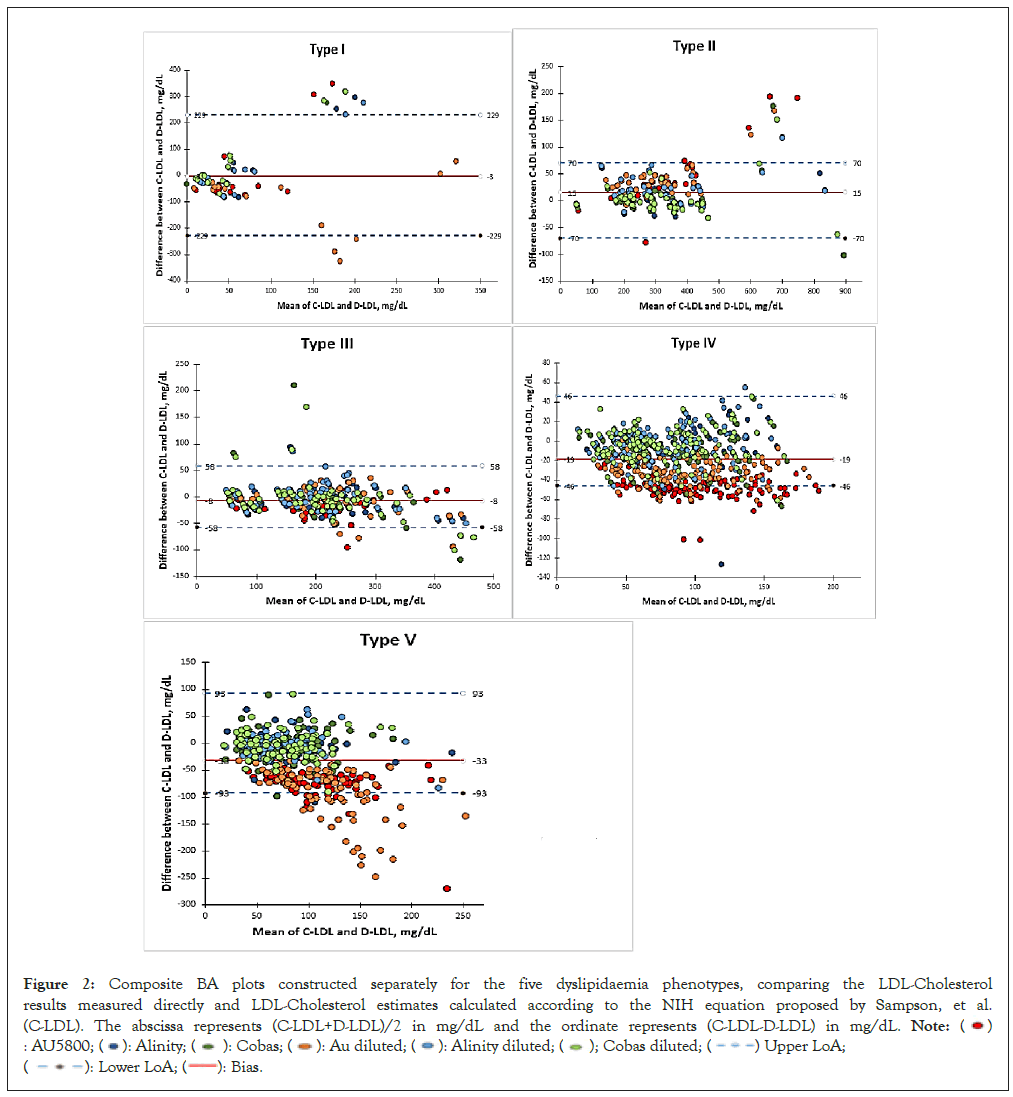
Figure 2: Composite BA plots constructed separately for the five dyslipidaemia phenotypes, comparing the LDL-Cholesterol
results measured directly and LDL-Cholesterol estimates calculated according to the NIH equation proposed by Sampson, et al.
(C-LDL). The abscissa represents (C-LDL+D-LDL)/2 in mg/dL and the ordinate represents (C-LDL-D-LDL) in mg/dL. 

For Type V specimens, the Bias is even greater (-33 mg/dL) and a clear dichotomy is noticed in the distribution of the points: most of the AU5800 results on diluted and undiluted specimens fall on the negative side of the bias line while most of the Alinity ci and Cobas Pure results fall on the opposite side, though evenly distributed above and below the zero ordinate. Thus, for Type V specimens too, AU5800 overestimates LDL-C levels and as the BA plot indicates the overestimation increases after dilution of the specimens.
BA plots comprehensively highlighted the role of each platform in contributing to the variation in D-LDL results in each of the dyslipidaemia phenotypes. However, quantification of the variation caused by each instrument platform within each dyslipidaemia phenotype needed to be enumerated. Herein lay the importance of ROC curves.
Using the NCEP (1995) guidelines [13], all D-LDL results with <12% of variation vis-á-vis C-LDL estimates were designated as ‘Concordant’ and those with variation of 12% and above were designated as ‘Discordant’. ROC curves were constructed for each of the five dyslipidaemia phenotypes with multiple comparator functions for the six data series as described in the materials and methods section with 1-specificity (False positive rate) as the abscissa and sensitivity (True positive rate) as the ordinate. The six data series in each panel represent LDL-Cholesterol estimations on the undiluted specimens performed on the three platforms AU5800, Alinity ci and Cobas Pure; and those on the same specimens diluted with normal saline and retested on the same three platforms (Figure 3).
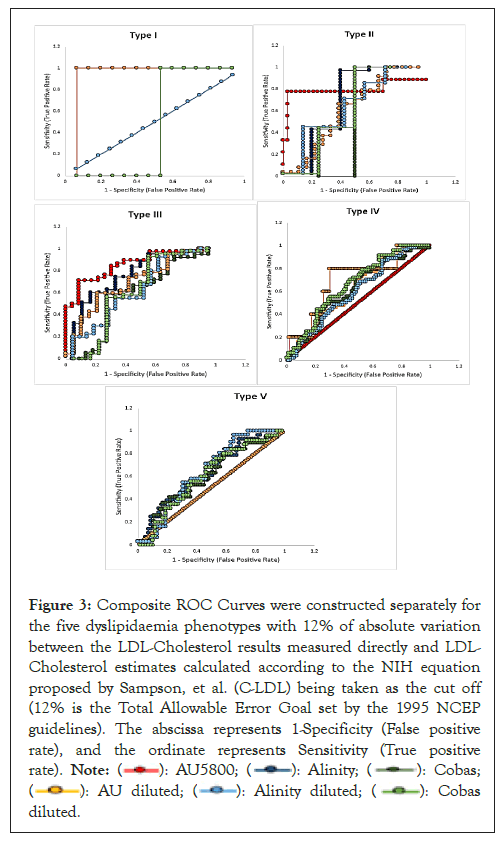
Figure 3: Composite ROC Curves were constructed separately for
the five dyslipidaemia phenotypes with 12% of absolute variation
between the LDL-Cholesterol results measured directly and LDL-
Cholesterol estimates calculated according to the NIH equation
proposed by Sampson, et al. (C-LDL) being taken as the cut off
(12% is the Total Allowable Error Goal set by the 1995 NCEP
guidelines). The abscissa represents 1-Specificity (False positive
rate), and the ordinate represents Sensitivity (True positive
rate). 

For Type I specimens, the curves representing AU5800, Alinity, Cobas and Alinity Diluted coincided with each other with 100% discordant results. Though the curve for AU Diluted exhibited maximum sensitivity (Area Under Curve (AUC 0.867)) and that of Cobas Diluted showed least sensitivity (AUC 0.4), these data should be cautiously interpreted due to the paucity of Type I specimens. For Types II and III specimens, AU5800 data exhibited maximum sensitivity (AUC 0.747 and 0.812 respectively), while Alinity and Cobas series of data also showed significantly improved sensitivity; for none of the three series, dilution helped in improving their respective AUCs. For Type IV specimens, AU5800 data exhibited 100% discordant results; dilution helped improve the sensitivity of AU5800 results (AUC 0.689); results on Alinity ci for the diluted and undiluted specimens coincided with each other proving dilution as a redundant technique for improving its results; for Cobas Pure, dilution marginally improved its AUC. In case of Type V specimens, both AU5800 and AU Diluted series exhibited 100% discordant results, thereby showing that dilution doesn’t help improve AU5800 results beyond 700 mg/dL of triglycerides; the other four series of data almost mirror each other and are marginally better in sensitivity than AU5800 (Table 4).
| Data set | AU5800 | Alinity | Cobas | AU diluted | Alinity diluted | Cobas diluted |
|---|---|---|---|---|---|---|
| Type I | 0.437 | 0.437 | 0.533 | 0.867 | 0.437 | 0.4 |
| Type II | 0.747 | 0.513 | 0.75 | 0.556 | 0.502 | 0.631 |
| Type III | 0.812 | 0.705 | 0.535 | 0.643 | 0.575 | 0.55 |
| Type IV | 0.499 | 0.607 | 0.613 | 0.689 | 0.582 | 0.648 |
| Type V | 0.488 | 0.616 | 0.59 | 0.488 | 0.62 | 0.587 |
Note: AU5800 represents results of undiluted samples on AU5800; Alinity represents results of undiluted samples on Alinity ci; Cobas represents results of undiluted samples on Cobas Pure; AU diluted represents results of diluted samples on AU5800; Alinity diluted represents results of diluted samples on Alinity ci and Cobas diluted represents results of diluted samples on Cobas pure.
Table 4: Area under curve statistics of the receiver operating characteristics curves.
As has been abundantly demonstrated in the Results section, there is wide variation in results of D-LDL between the three platforms tested across all the dyslipidaemia phenotypes, but most prominently when the Triglycerides concentrations are high, viz Types I, IV and V. Even in Types II and III, there is variation in D-LDL results between the platforms in extremes of Non-HDL Cholesterol concentrations. In the absence of credible comparison studies against reference methods, the dilemma arises as to which platform should be used for reporting dyslipidaemic specimens. Though the present study does not validate its results directly against any reference method, it compares its results with the C-LDL values derived from the NIH equation proposed by Sampson et al, which is itself validated against β-quantification. The results of this comparison study put the AU5800 platform directly into focus both for its advantages, as well as its disadvantages when it comes to D-LDL estimation. While its performance in estimating D-LDL concentrations in Type II and III specimens is commendable, generating AUCs of 0.747 and 0.812 respectively on the ROC curves, its performance is dismal in estimating D-LDL concentrations in Types I, IV and V specimens, generating 100% discordant results. The results of the analysis indicate that AU5800 significantly overestimates D-LDL results in presence of high Triglycerides concentrations. On the other hand, Alinity ci and Cobas Pure consistently deliver moderately concordant results across all the dyslipidaemia phenotypes, but even their performance dips significantly in case of lipaemic specimens. Dilution and retesting of the specimens does not help improve the results in the AU5800; in fact, the variation worsens in Type V, as evident from the BA plot, though the AUC in the corresponding ROC curve improves marginally. Dilution does help improve the AUC counts of the other two platforms, but only marginally.
The implications of the above findings are manifold. It was partly due to the inaccuracy of D-LDL results that guidelines have come up emphasizing on non-HDL-C as the main biomarker for monitoring of hypertriglyceridaemic subjects, for example the American College of Cardiology and American Heart Association guidelines [14]. However, reporting of D-LDL results cannot be completely done away with, as more recent guidelines from the same entities have again put emphasis on the LDL-C results [15]. Moreover, accurate determination of LDL-C is imperative for precise characterization of subjects with high triglyceride-rich lipoprotein cholesterol, in view of large population-based studies implicating the latter to be more atherogenic than the former [16,17]. In view of the above concerns, the present author is of the considered opinion that harmonization of different platforms for estimation of D-LDL in dyslipidaemic specimens, is a task which is long overdue.
This study is not devoid of limitations and its author duly recognizes the fact. The most obvious limitation, as mentioned earlier, is the absence of a direct comparison with any reference method; to overcome this shortcoming, results were compared to calculated LDL-C by a method which was itself validated against β-quantification. The second limitation of this study is the low number of subjects. Agreed that the inclusion criteria for the study was quite demanding, this study was especially hamstrung when it came to analysis of Type I results. For the other dyslipidaemia phenotypes, the conclusions were quite unambiguous, but the author acknowledges that results would have been more acceptable for Type I had its sample size been larger. Thirdly, dyslipidaemia phenotypes were determined based on only clinical history, findings of clinical examination and pattern of standard lipid profile and not on the basis of genotype profiling or use of specialized markers like Apolipoprotein A1, Apolipoprotein B and Lipoprotein (a). The author acknowledges that use of at least the specialized markers would have been an improvement. Fourthly, this study excludes subjects who are on lipid lowering drugs. While this strategy simplifies the protocol of the study by minimizing variables, it sacrifices the findings on a crucial group of subjects which may be important for clinical investigators. Finally, this study included only fasting lipid profile results, again, for simplifying the study protocol and as a result, missed out on crucial information regarding post-prandial changes in D-LDL estimation.
This study shows that when it comes to dyslipidaemic subjects, results of directly measured LDL-Cholesterol on three popular platforms viz AU5800, Alinity ci and Cobas Pure vary widely enough to cause misclassifications during diagnosis or monitoring of treatment. When compared to calculated LDL-Cholesterol derived from the NIH equation proposed by Sampson et al, results of directly measured LDL-Cholesterol on AU5800 correlate well in case of Type II and III subjects, but is overestimated in case of Type I, IV and V subjects; those on Alinity ci and Cobas Pure correlate moderately in case of Types II, III and IV subjects but poorly in case of Types I and V subjects. Detailed study of these implications needs to be undertaken for the betterment of patient care outcomes.
The author declares that there has been no conflict of interests with any individual or organization in the conduct of this study and further declares that no funding has been received from anywhere for this study.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Sarkar R (2023) Variability in Directly Measured LDL-Cholesterol in Dyslipidaemic Specimens: Is there a Cause for Concern? J Clin Chem Lab Med. 6:275.
Received: 16-Nov-2023, Manuscript No. JCCLM-23-28040; Editor assigned: 20-Nov-2023, Pre QC No. JCCLM-23-28040 (PQ); Reviewed: 04-Dec-2023, QC No. JCCLM-23-28040; Revised: 11-Dec-2023, Manuscript No. JCCLM-23-28040 (R); Published: 18-Dec-2023 , DOI: 10.35248/2736-6588.23.6.275
Copyright: © 2023 Sarkar R. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.