Virology & Mycology
Open Access
ISSN: 2161-0517
ISSN: 2161-0517
Research Article - (2020)Volume 9, Issue 1
Background: The remarkable diversity and mobility of Newcastle disease viruses (NDV) includes virulent viruses of genotype VI. These viruses are often referred to as pigeon paramyxoviruses 1 because they are normally isolated and cause clinical disease in birds from the Columbidae family. Genotype VI viruses occasionally infect, and may also cause clinical disease in poultry. Thus, the evolution, current spread and detection of NDV are relevant to avian health.
Methods: Nucleotide sequencing and phylogenetic studies of three Kazakhstan isolations were performed to characterize the complete fusion (F)-protein gene as well as whole genome sequence. Sequence data were compared with 106 Fusion genes representing different NDV genotypes and sub-genotypes, as well as 225 fusion genes and 37 whole genome of class VI NDV strains from different regions of the world at different time periods. Phylogenetic trees were constructed to determine evolutionary relationships among these strains. We analysed fusion (F) protein gene and whole genome sequences, including the cleavage site.
Results: The complete genome of these 3 isolates contained 15142bp, 15085bp and 15102bp in length, similar to those of Newcastle disease virus (NDV) strains in genotypes VIg, with the gene order 3’-NP-P-M-F-HN-L-5’. The cleavage site of the fusion protein was 112KRQKR116-F117, a feature generally associated with virulent NDV strains. Phylogenetic analysis, based on genomic sequences, SNP and fusion gene sequences, revealed that three isolates should be classified as class II genotype VIg NDVs. Phylogenetic analysis revealed that the NDV from various sites in Kazakhstan was highly similar genetically and that it clustered together with NDV of genotype VIg. Based on our data analysis, VIg isolates shared highest sequence identity with Russian and Ukraine isolates of the VIg subgenotype, suggests the possible spread of velogenic NDV in this region through cross-border live bird trade.
Conclusion: Our study provides baseline information on the genetic characteristics of NDV circulating in Kazakhstan and we propose that the evolutionary and epidemiological study of virulent NDV could help to provide accurate molecular data about variants circulating in this region, thus aiding in the design of more efficient recombinant vaccines.
Newcastle disease virus; Avian orthoavulavirus 1; Wild birds; Kazakhstan
Newcastle disease (ND) is a highly contagious viral infection in domesticated and wild birds occurring worldwide. ND, alongside with avian influenza, is considered one of the two most devastating diseases in wild birds and poultry industry. It has become endemic in many areas, such as in Kazakhstan since its first recorded outbreak in 1980, resulting severe economic consequences. ND is caused by virulent strains of Newcastle disease virus (NDV), commonly known as avian paramyxovirus 1, which was previously designated as Avian avulavirus 1. The International Committee on Taxonomy of Viruses has recently created three genera, named Orthoavulavirus, Metaavulavirus, and Paraavulavirus, within a new subfamily Avulavirinae of the family Paramyxoviridae. Viruses of genus Avian orthoavulavirus 1(AOAV-1) (formerly designated as Avian avulavirus 1 (AAvV-1)) was commonly knownas Avian paramyxoviruses 1 (APMV-1) or Newcastle disease viruses (NDV) - ICTV 2019 (https://talk.ictvonline.org/taxonomy/) [1-3].
Of these, AOAV-1, synonymous with Newcastle disease virus (NDV), is the most widely characterized and studied due to the economic importance of Newcastle disease (ND) caused by virulent strains of the virus. NDV has a single stranded, non-segmented, negative-sense RNA genome consisting of six genes in order of 3’to 5’: nucleocapsid (NP), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin-neuraminidase (HN), and polymerase (L), coding for these six structural proteins and at least one additional V protein [4,5].
Newcastle disease viruses (NDV) are genetically grouped into two divergent classes that are further classified in genotypes and subgenotypes [6-8]. Transmission between host species appears to be an important 82 mechanism contributing to the maintenance, propagation, and spread of NDV.
Bi-directional viral exchange between domestic and wild bird populations has also been indicated by routes such as viral diffusion. Previous research has shown the presence of live vaccine strains intended for domestic poultry in wild birds [9,10]. It has been known that NDV genotype VIg can be transmitted from pigeons and doves to domesticated chicken especially at ecological contact surfaces [11,12], and that some of them dramatically gain virulence upon a few passages in chicken [13,14].
Their occurrence in pigeon population economically threatens the poultry subsector in the region. Kazakhstan is on the main pathways of transcontinental migratory routes of many wild birds. Genomic analysis of circulating NDV strains in Kazakhstan can potentially provide insights to NDV genetic evolution and pathogenic characteristics. Currently, only limited studies on the genetic variability of NDV strains in Kazakhstan have been conducted, and few NDV whole genome sequences are available.
Newcastle disease viruses are constantly evolving and different genetic groups undergo simultaneous evolutionary changes in different geographical locations making the available genetic makeup information outdated [5,15]. These evolutionary changes present challenges to prompt diagnosis. Some currently validated methods are target-oriented and might fail to detect new viral genetic variants [16-18].
Lack of comprehensive NDV genetic information further hinders understanding of Newcastle disease evolution and epidemiology. Furthermore, mixed viral infection is not uncommon in animals, and in the case of poultry, they are quite frequent [19]. Efficient and accurate detection of these pathogens is important for the creation of appropriate strategies for disease control and disease
106 surveillance in live birds, households and commercial poultry farms [20]. Full-length viral genome sequencing allows for de novo, high-quality genetic characterization of NDV. In this study, we selected three NDV isolates from Kazakhstan region and performed whole genome sequencing. Partial sequences of these three NDV strains have been previously generated and deposited into GenBank (NDV 15 submission # 2294838NDV 7, Submission # 2314571, NDV 10 Submission # 2314574).
Sample collection
A large number of tracheal and cloacal samples were collected for wild bird viral surveillance monitoring in 2011 and 2014, in the Shakpak Ornithological Station in the Zambyl Oblast and other regions in the Republic of Kazakhstan. The three selected NDV isolates for this study were from 2014 collection of tracheal swabs [21]. Fulica atra/KZ/WKO/4/2014 was collected from the Eurasian coot (Fulica atra) in the West KZ Oblast; Acridotheres/KZ/ Zhambyl/17/2014 from common myna (Acridotheres tristis) and Columba livia/KZ/Zhambyl/32/2014 from rock dove (Columba livia), both in Zambyl Oblast.
Viral RNA extraction
This study used the previously prepared virus isolation from specific-pathogen-free embryonated chicken eggs [21]. Lyophilized samples were diluted in the original volume with water (1 ml). The suspension was used for RNA extraction using the QIAamp Viral RNA Mini Kit (Qiagen, UK), according to the manufacturer's instructions. RNA Elution was performed using water, 2 times, 40μl in each case.
Genome sequencing
A set of primers was designed to target NDV genotype VI. A subset of full NDV genome sequences from GenBank were selected as input to the NCBI Primer-BLAST. Considerations to optimize the primers included primer length, GC content and melting temperature. The primers were commercially synthesized, and diluted to a working concentration of 10 pmol. The PCR was performed using a SuperScript III One-Step RT-PCR System Kit (ThermoFisher Scientific). 24 pairs of primers were used for the VI genotype (Table S1) thus preparing a master mix for 25 reactions. Purification of PCR products was carried out using the AMPure kit (Beckman Culter), according to the manufacturer's instructions. The quality of the PCR products was checked on electrophoretic analysis. PCR products for sequencing were obtained with the BigDye® Terminator v3.1 Cycle Sequencing Kit. Purification of the sequencing reaction was carried out using the Clean Seq kit. Sequencing was performed on a 16 capillary 3130Xl Genetic Analyzer sequencer (Applied Biosystems/Hitachi). Genomic assembly, genome annotation, comparative genomics, and phylogenetic analysis were carried out using the CLC Genomic Workbench 12 program.
Phylogenetic analysis
Whole genome tree: The three newly sequenced NDV genome sequences combined with 37 complete genomes of genotype VI NDV downloaded from GenBank were used for complete genomebased analysis. Phylogenetic trees were constructed by Maximum Likelihood Phylogeny 1.3 on CLC Genomics Workbench 12, using the ‘Neighbour Joining’ method and the Jukes Cantor model with Gamma distribution parameter 1.0 and 100 bootstrap replications to assign confidence levels to branches.
SNP tree analysis: The same 37 available complete genomes of NDV in genotype VI were used for SNP based analysis. The SNP tree was generated by using PhaME-1 version 1(https://github.com/ mshakya/PhaME-1) with default settings. The phylogenetic trees were inferred with RAxML-Best tree and bootstrap valve mapped on Bipartitions tree. F gene tree analysis: Reference NDV genomes listed in the Table S1 of Ogali, et al. were selected and downloaded from GenBank (n=106) [22]. To represent all NDV genotypes. At least three sequences representing each genotype were used in the analysis. All known NDV isolated from KZ deposited in GenBank are downloaded. There are total 76 NDV strains isolated in Kazakhstan, 106 previously described NDV sequences representing different NDV genotype groups were used for comparison. Multiple alignment and comparison of three study sequences and downloaded GenBank sequences were performed using MUSCLE v3.8.31 [23]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) version 7.0 [24]. We constructed phylogenetic trees using the maximum likelihood (ML) method and estimated the tree using the Kimura 2-parameter model of nucleotide substitution with gamma-distributed rate variation among sites. We employed a bootstrap resampling process (100 replications) to assess the robustness of individual nodes of phylogeny. For building VI group subtree, all sequences belong to VI group in the Table S1 of Dimitrov, et al. were downloaded from GenBank and curated resulting in a large datasets (n =225) [8]. Together with three sequences obtained in the current study, all sequences were aligned and constructed a phylogenetic tree according methods descripted above (Table 1).
| Name | Sampling place | Date | Bird species |
|---|---|---|---|
| Fulica atra/KZ/WKO/4/2014 | Aktobe region, Martuk district, Bulak Lake | Apr. 2014 | Eurasian coot |
| Columbalivia/KZ/Zhambyl/32/2014 | Zhambyl, Shakpak ornithological station | Oct. 2014 | Rock dove |
| Acridotheres/KZ/Zhambyl/17/2014 | Zhambyl, Shakpak ornithological station | Oct. 2014 | Common myna |
Table 1: Strains isolated from the samples.
Genbank accession number
The nucleotide sequences determined in this study are available in the GenBank under accession numbers
Acridotheres/KZ/Zhambyl/17/2014 (NDV-15) MT081320
Columbalivia /KZ/Zhambyl/32/2014 (NDV-7) MT081321
Fulica atra/KZ/WKO/4/2014 (NDV-10) MT081322
Geographic sampling location
Three isolates were selected from positive samples from a previous study [21]. These three isolates that had been previously sequenced its fusion gene, were selected from the Research Institute of Biological Safety Problems (RIBSP) collection for whole genome sequencing and analysis. These samples collection sites are shown in Figure 1.
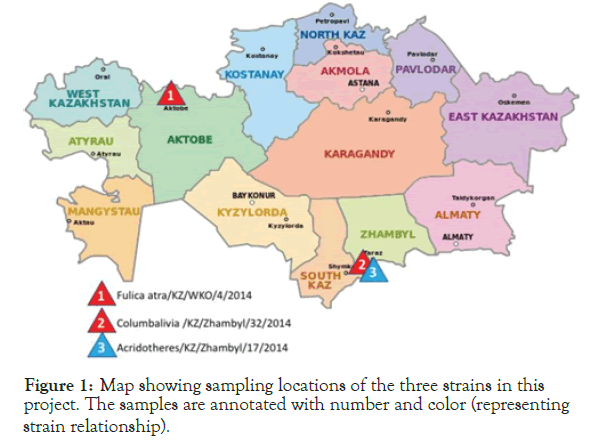
Figure 1. Map showing sampling locations of the three strains in this project. The samples are annotated with number and color (representing strain relationship).
Genome length
Whole genome assembly reveals that genome sizes for three Kazakhstan ND strains Fulica atra/KZ/WKO/4/2014, Columbalivia/KZ/Zhambyl/32/2014, and Acridotheres/KZ/ Zhambyl/17/2014 are 15142bp, 15085bp and 15102bp respectively. The length variation occurs at the upstream UTR region of nucleocapsid protein (NP) gene (Table 1).
Coding regions
Similar to other NDV strains, the genome of these three isolates contained six genes in the order of 3'-NP-P-M-F-HN-L-5' as expected. The protein coding capacity of the genome of these three isolates was the same at approximately 90.6%. The genomic features and protein characteristics of three isolates are summarized in Table 2.
| Gene | Gene length (nt) | ORF (nt) | 3' UTR (nt) | 5' UTR (nt) | Intergenic region (nt) | Amino acid length (aa) |
|---|---|---|---|---|---|---|
| NP | 1752 | 1470 | 66 | 216 | 2 | 489 |
| P | 1451 | 1188 | 83 | 180 | 1 | 395 |
| M | 1241 | 1095 | 34 | 112 | 1 | 364 |
| F | 1792 | 1662 | 46 | 84 | 31 | 553 |
| HN | 2002 | 1716 | 91 | 195 | 47 | 571 |
| L | 6703 | 6615 | 11 | 77 | 2204 |
Table 2: Genetic characteristics of three Kazakh NDV strains.
Non-coding regions
The AOAV-1 genome started from the 3′ -leader region, and the genes were transcribed into separate mRNAs through a start–stop– restart mechanism guided by the conserved gene start (GS) and gene end (GE) of the genes. Located between GS and GE were the non-coding intergenic sequences (IGS), which ranged from 1 nt to 47 nt. The open reading frame of each gene overhanged with the 3' and 5' untranslated regions (UTRs) on their respective ends. The sequences of the GS and GE as well as of the IGS in 3 strains are summarized in Table 2. The GS for the five NP, P, M, F and HN genes was ACGGGTAGAA, whereas that for the L gene was ACGGGTAGGA. The GE for NP, M and L was TTAGAAAAAA, whereas that for P, HN and F gene was TAAGAAAAAA. The lengths of intergenic sequence (IGS) of P-M and M-F were 1 nt, while the IGS of NP-P, F-HN and HN-L were 2 nt, 31 nt and 47 nt, respectively (Table 3).
| NDV gene | Percent identity | Number of amino acid substitutions |
|---|---|---|
| Nucleocapsid Protein (NP) | 98.8 | 6 |
| Phosphoprotein (P) | 95.1 | 19 |
| Matrix Protein (M) | 99.7 | 1 |
| Fusion Protein (F) | 98.4 | 9 |
| Hemagglutin Neuraminidase (HN) | 96.8 | 18 |
| Large Polymerase Protein (L) | 98.4 | 17 |
Table 3: Amino acid sequence comparison of Fulica atra/KZ/WKO/4/2014 and Acridotheres/KZ/Zhambyl/17/2014.
Comparative study
A comparative analysis of the complete genomes of the strain Fulica atra/KZ/WKO/4/2014 and Acridotheres/KZ/Zhambyl/17/2014 reveal 99% coverage and 96.4% identity between them. The genome comparison reveals the Columbalivia/KZ/Zhambyl/32/2014 is identical to Fulica atra/KZ/WKO/4/2014, except for one nucleotide substitution in the hemagglutinin-neuraminidase (HN) gene and 57bp longer at the upstream UTP region of NP gene in Fulica atra/KZ/WKO/4/2014.
Megablast against GenBank revealed these 3 strains having 97% identity with strains isolated in Russia in 2009–2011 and 95% identity with a strain isolated in Ukraine in 2013. The pairwise comparison among 3 strains, NDV/Altai/pigeon/770/2011, Pi/Rus/Kemerovo/0267/09, and pigeon/Ukraine/ Doneck/3/968/2007 reveal that NP and M genes were the most conserved with sequence identities that ranged from 97.34% to 99.73% (nucleotide) and 1 to 13 (amino acid changes) in all strains. In contrast, the P-gene was the most variable with sequence identities that ranged from 91.92% to 96.46% (nucleotide) and 14 to 32 (amino acid changes), depending on the strain. Analysis of sequence identities using available complete genome sequences from the GenBank showed that Altai/pigeon/777/2010, Pi/ Rus/Kemerovo/0267/09, and Altai/pigeon/777/2010 were most closely related to NDV- 10(97.18%), NDV-15 (97.23%), and NDV-7 (97.18%) respectively.
F gene comparison
Analysis of the primary sequences of the fusion proteins (F protein) revealed that all strains exhibited the cleavage site sequence 112KRQKR116- F117, which is typical for pathogenic strains of NDV. Analysis of the functional domains of the F-gene sequence of three strains showed that the deduced amino acid sequences were mostly similar with the consensus amino acid sequences derived from 37 NDV strains in genotype VI; except that the F protein of Fulica atra/KZ/WKO/4/2014 and Acridotheres/KZ/ Zhambyl/17/2014 had 0/1 at the transmembrane domain, 3/1 at the signal peptide, and 0/0 at the fusion peptide respectively. Analysis of the three heptad repeat regions (HR) showed a total of 1/0 substitutions in HRa (143–185 amino acids), and 2/2 substitutions in HRc (471–500 amino acids) respectively (Table 4).
| Strains | Signal peptide (1-31) | Fusion peptide (117-141) | Hra (143-185) | HRc (471-500) | Transmembrane domain (501-521) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Consensus | 2 | 13 | 22 | 125 | 146 | 476 | 485 | 490 | 499 | 509 |
| G | L | I | V | Q | N | N | K | S | V | |
| Fulica atra/KZ/WKO/4/2014 | S | P | T | V | R | S | N | K | P | V |
| Columbalivia/KZ/Zhambyl/32/2014 | S | P | T | V | R | S | N | K | P | V |
| Acridotheres/KZ/Zhambyl/17/2014 | S | L | I | V | Q | S | N | R | S | A |
| Pi/Rus/Kemerovo/0267/2009 | S | L | I | V | Q | S | S | K | S | A |
| Altai/pigeon/770/2011 | S | L | I | I | Q | S | N | K | P | V |
Table 4: Amino acid substitutions in the functional domains of the F protein.
Phylogenetic analysis
We illustrate the relationship between all 106 Kazakh isolates deposited in Genbank plus 3 Kazakh strains obtained in this study and other 76 NDVs available in GenBank, using a phylogenetic tree of F gene sequence of the Kazakh isolates obtained in the study and the other GenBank sequences of NDV (Figure 2).
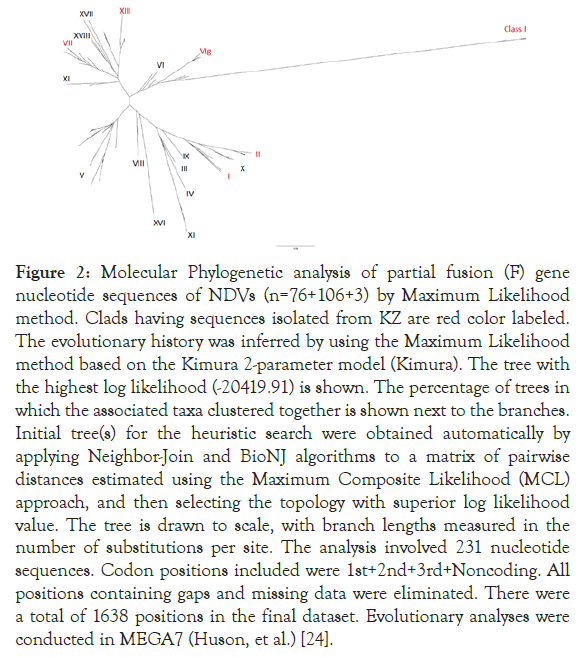
Figure 2. Molecular Phylogenetic analysis of partial fusion (F) gene nucleotide sequences of NDVs (n=76+106+3) by Maximum Likelihood method. Clads having sequences isolated from KZ are red color labeled. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model (Kimura). The tree with the highest log likelihood (-20419.91) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 231 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1638 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Huson, et al.) [24].
From the 241 tree, all three sequences were grouped together with other sub-genotype VIg sequences from KZ. Zoom-in subgenotype VIg clad, Columbalivia/KZ/Zhambyl/32/2014 and Fulica atra/ KZ/WKO/4/2014 were more closely identical (Figures 2 and 3) to previous KZ strains (MK693035.1_AAvV 1/rock_dove/ Southern_Kazakhstan/6444/2014; MK693038.1_AAvV-1/ golden_eagle/Southern_Kazakhstan/6451/2014; KT965728.1 columbalivia/KZ/Zhambyl/32/2014; KT965731.1 buteo rufinus/ KZ/Zhambyl/41/2014;KT965730.1 anas acuta/KZ/EKO/1/2014) sequences.
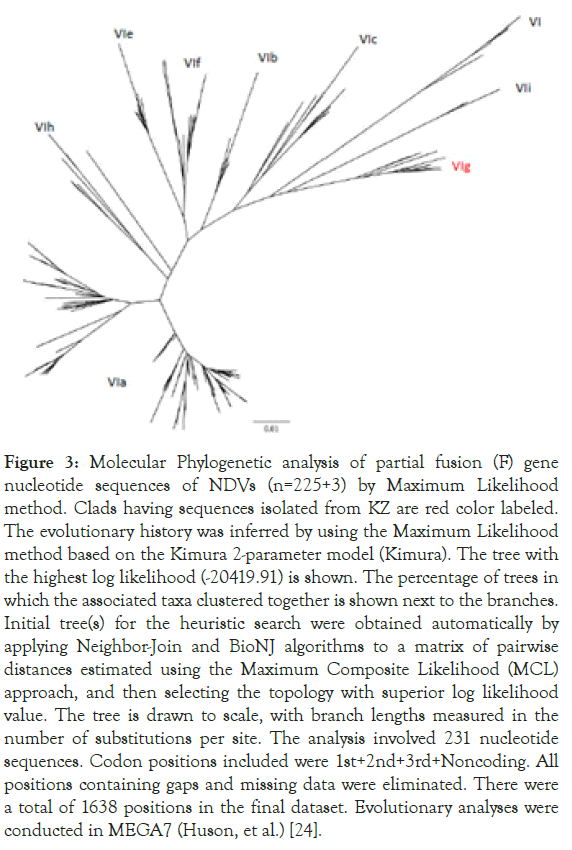
Figure 3. Molecular Phylogenetic analysis of partial fusion (F) gene nucleotide sequences of NDVs (n=225+3) by Maximum Likelihood method. Clads having sequences isolated from KZ are red color labeled. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model (Kimura). The tree with the highest log likelihood (-20419.91) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 231 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1638 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Huson, et al.) [24].
While Acridotheres/KZ/Zhambyl/17/2014 was more closely identical to previous KZ strains (JN806237.1 AAvV- 1/ pigeon/Almaty/1065/05; KT965732.1 columbalivia/KZ/ Zhambyl/27/2014; and KT965729.1 acridotheres/KZ/ Zhambyl/17/2014) sequences.
This study VIg sequences together with previous KZ strains formed a distinct clade branching with 100% bootstrap value at the defining node from other subgenotypes of genotype VIg including genotype VIg viruses from Europe and Africa, such as JF824032.1 Pi/Rus/ Vladimir/687/05, JQ039385.1 dove/Nigeria/VRD07163/2007 and JF824013.1 Pi/Rus/Kemerovo/0267/09.
By checking with the latest NDV classification and nomenclature system proposed by international consortium, F gene of Columbalivia/KZ/Zhambyl/32/2014/Fulica atra/KZ/ WKO/4/2014 (NDV-7 and NDV-10) and Acridotheres/KZ/ Zhambyl/17/2014 (NDV-15) are identified as XXI.1.1_VI_g_735_ KT965728_pigeon_Kazakhstan_Zhambyl_32_2014 and XXI.1.1_ VI_g_734_KT965727_pigeon_Kazakhstan_EKO_15_2014 respectively in the Supplemental Dataset S2 (class ii) [25].
Both sequences belong to new genotype XXI. In addition, a phylogenetic analysis of three Kazakh strains and 37 NDV genotype VI strains was conducted on the basis of the complete genome sequence using the neighbour-joining algorithm with bootstrap 500 replicates on CLC Genomic Workbench v12. All 3 isolates were grouped under class II NDVs in genotype VIg (Figure 4). 3 KZ strains phylogenetically close to other VIg strains, such as KT962979 Altai/pigeon/777/2010, KJ920204 NDV/Altai/ pigeon/770/2011, and JF827027 Pi/Rus/Kemerovo/0267/09 isolates. Phylogenetic analysis using whole genome SNP yielded the same tree topology and phylogenetic groupings (Figure 5).
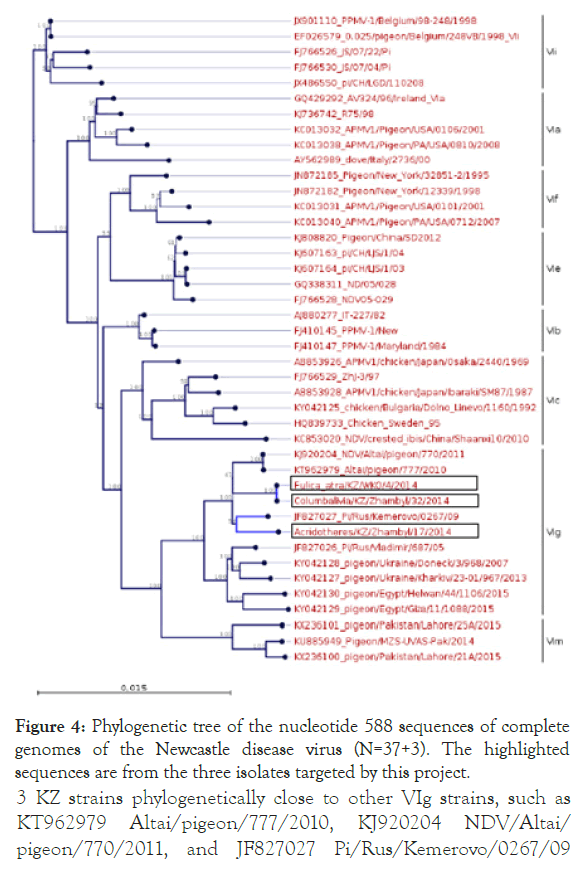
Figure 4. Phylogenetic tree of the nucleotide 588 sequences of complete genomes of the Newcastle disease virus (N=37+3). The highlighted sequences are from the three isolates targeted by this project.
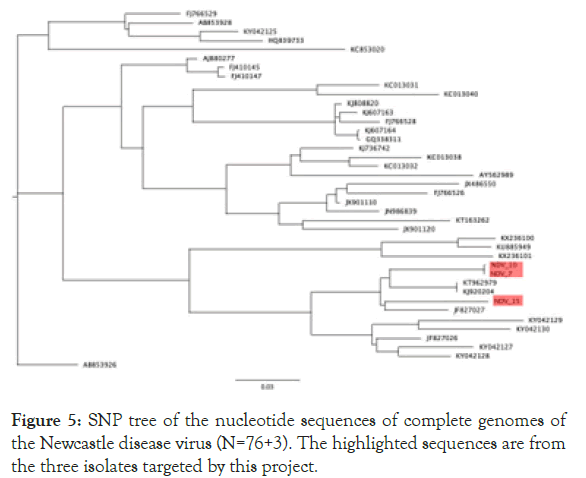
Figure 5. SNP tree of the nucleotide sequences of complete genomes of the Newcastle disease virus (N=76+3). The highlighted sequences are from the three isolates targeted by this project.
This study focused on molecular and genomic characterization of three NDV isolates from wild birds from a larger study in Kazakhstan. Phylogenetic analysis using 3+37 complete NDV genome sequences in genotype VI revealed that three Kazakh strains belong to subgenotype VIg. SNP Phylogenetic analysis using the same input yielded the same phylogenetic groupings. Subgenotype analyses using curated VI gene sequences showed that all three strains belong to the subgenotype VIg. This topology of the F-gene phylogenetic tree indicated the viruses from three strains isolated from KZ grouped together and clustered within rest of subgenotype VIg. These three viruses were closely related to viruses isolated from pigeons in Russia, Ukraine, and Kazakhstan during 2005–2014 [26-28].
These results were consistent with the results of the complete genome analysis, as well as updated unified phylogenetic classification system based on the complete fusion gene sequences of isolates representing NDV class II [25]. The new phylogenetic classification system has shown these three NDV isolates belonging to new genotype XXI Dimitrov, et al. together with other XXI sequences from Ukraine, Russia, Kazakhstan, as well as distanced regions from Egypt, Nigeria, and Pakistan [25]. As these kind of close genetic relationships among these isolates from different regions, it could be of epidemiological significance and certainly suggest a recent common ancestry during their evolution [29,30]. Multiple virulent subgenotype class I, class II-I, II, VIg, VII and XIII NDV virus have been showing circulating in KZ. It has been known that class II genotypes I, V, VI and VII have been isolated in many countries and avian species, suggesting viral dispersion across geographic areas and among diverse hosts [8,31]. Viruses from sub-genotype VIg also have been known being isolated from pigeons and a dove in Nigeria, Russia, and Ukraine during 2005–2011 Van Borm, et al., Pchelkina, et al., Yurchenko, et al. and Iran during 2012 to 2014 Mayahi, et al., as well as in different West and Central African countries (Snoeck, Adeyanju et al. 2013, Hicks, et al. [26-29,32,33]. An interesting observation from this study is that two of NDV virus samples from different bird species and oblasts (Aktobe oblast in Western KZ and Zhambyl oblast in South KZ) were identified as belonging to the same strain. These two specimens with nearly identical genomes originated from birds of different species separated by about 1500km. This illustrates how the virus can move rapidly between different species of wild birds over long distances. Such transitions from an established host into a new recipient host have been inferred to occur, as evidenced by host-adapted lineages: Columbiformes-adapted genotype VI viruses, (Hicks, et al. [33]. A possible explanation for the spread of these viruses over long distances is contact between columbid birds during competitive flights, exhibitions, or due to such birds 'intensive international trade [34]. Other possibilities are the international trade between countries of live birds from other species or avian products, either through legal or illegal import and export routes. Due to the close proximity of multiple bird species and the frequent turnover of naive hosts, these live bird markets can represent a distinct ecological niche that could generate minimal cross-species transmission for AOAV-1 [33].
In addition, NDV of genotype VI have been isolated from birds from non-Columbidae species kept in captivity and from wild birds, including perdrixes, faisans, swans, falcons, blackbirds, cockatoos, budgerigars, raptors, pertridges, crested ibises, waterfowl, starlings, pintails, gannets, and buzzards [35-38]. Genotype VI viruses have previously been reported to circulate in apparently healthy pigeons Teske, et al., Byarugaba, et al., Wang, et al. and recent work on the VI-restricted model of genotype Hicks, et al., and a previously published phylodynamic study of genotype VI Chong, et al., showed that global migration of VI genotype could be driven by pigeon transport, resulting in viral dispersal from Europe to other regions, including Africa and East Asia [33,39-41].
Because live animal movement (i.e., trade and wild bird migration) are the most likely drivers of global AOAV-1 spread, inanimate, vaccine and non-avian animal contamination may also serve as mechanisms for viral migration Yates, et al., Alexander, et al., Alexander, et al., Jorgensen, et al., Li, et al., Sharma, et al., Pedersen, et al., Zhao, et al., these could explain why major poultry trade partners are also reflected in the global diffusion network [42-49].
Specifically, Europe, which is a net exporter of poultry to the rest of the world Bahl, et al., was supported as an important source of class II AOAV-1 [50]. The appearance of fecal and oral swabs of genotype VI viruses indicates viral replication, which could result in virus transmission and possible outbreaks in poultry, as seen previously Hicks, et al., is occurring in pigeons [33].
Although sporadic, NDV outbreaks in poultry caused by viruses of genotype VI have been reported, and the potential of the virus to cause clinical disease in poultry must not be under estimated, as NDV can be transmitted to poultry via wild birds; therefore, studying the strain variance in wild birds will give an indication of how to design diagnostic tests for poultry and enable improved readiness for any future outbreaks [43,51-54]. Especially considering countries in Africa, Europe and the Middle East are particularly underrepresented, continued surveillance and sampling among regions with low representation would improve future analyses and may be showing more important in the global diffusion of AOAV-1 than what have reported.
The results of this study also could be helpful for developing efficient recombinant vaccines that are homologous to viruses that are circulating in the country.
( h t tp://www. o i e . i n t/wahi s_2/publ i c/wahi d .php/ Diseaseinformation/Diseasetimelines).
This work was funded by the US Defense Threat Reduction Agency Biological Threat Reduction Program. Los Alamos National Laboratory, an affirmative action/equal opportunity employer, is managed by Triad National Security, LLC, for the National Nuclear Security Administration of the US Department of Energy under contract 89233218CNA000001. We thank T. Erkkila, D. Dasgupta, and MRIGlobal for assistance with the project implementation, and Dr. Claudio Afonso (formerly USDA) and Ms. Hajnalka Daligault (LANL) for assistance in primer design.
Citation: Strochkov V, Burashev Y, Sandybayev N, Xie G, Erkkila TH, Cui H, et al. (2020) Whole Genomes of Avian orthoavulavirus 1 (Newcastle Disease Virus Genotypes VIg or new genotype XXI.) in Wild Birds in Kazakhstan. Virol Mycol. 9:183. DOI: 10.35248/2161-0517.20.09.183.
Received: 03-Feb-2020 Accepted: 17-Feb-2020 Published: 24-Feb-2020 , DOI: 10.35248/2161-0517.20.09.183
Copyright: © Strochkov V, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.